Abstract
We review studies from our laboratories using different molecular tools to characterize the ancestry of Brazilians in reference to their Amerindian, European and African roots. Initially we used uniparental DNA markers to investigate the contribution of distinct Y chromosome and mitochondrial DNA lineages to present-day populations. High levels of genetic admixture and strong directional mating between European males and Amerindian and African females were unraveled. We next analyzed different types of biparental autosomal polymorphisms. Especially useful was a set of 40 insertion-deletion polymorphisms (indels) that when studied worldwide proved exquisitely sensitive in discriminating between Amerindians, Europeans and Sub-Saharan Africans. When applied to the study of Brazilians these markers confirmed extensive genomic admixture, but also demonstrated a strong imprint of the massive European immigration wave in the 19th and 20th centuries. The high individual ancestral variability observed suggests that each Brazilian has a singular proportion of Amerindian, European and African ancestries in his mosaic genome. In Brazil, one cannot predict the color of persons from their genomic ancestry nor the opposite. Brazilians should be assessed on a personal basis, as 190 million human beings, and not as members of color groups.
DNA tests; Insertion-deletion polymorphisms; Ancestry; Human diversity; Brazilians
Braz J Med Biol Res, October 2009, Volume 42(10) 870-876
DNA tests probe the genomic ancestry of Brazilians
 Correspondence and Footnotes
Correspondence and Footnotes
 S.D.J. Pena1,2, L. Bastos-Rodrigues2, J.R. Pimenta1 and S.P. Bydlowski3
S.D.J. Pena1,2, L. Bastos-Rodrigues2, J.R. Pimenta1 and S.P. Bydlowski3
1GENE, Núcleo de Genética Médica, Belo Horizonte, MG, Brasil
2Departamento de Bioquímica e Imunologia, Universidade Federal de Minas Gerais, Belo Horizonte, MG, Brasil
3Laboratório de Genética e Hematologia Molecular (LIM-31), Faculdade de Medicina, Universidade de São Paulo, Hospital das Clínicas, São Paulo, SP, Brasil
 References
References
 Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Abstract
We review studies from our laboratories using different molecular tools to characterize the ancestry of Brazilians in reference to their Amerindian, European and African roots. Initially we used uniparental DNA markers to investigate the contribution of distinct Y chromosome and mitochondrial DNA lineages to present-day populations. High levels of genetic admixture and strong directional mating between European males and Amerindian and African females were unraveled. We next analyzed different types of biparental autosomal polymorphisms. Especially useful was a set of 40 insertion-deletion polymorphisms (indels) that when studied worldwide proved exquisitely sensitive in discriminating between Amerindians, Europeans and Sub-Saharan Africans. When applied to the study of Brazilians these markers confirmed extensive genomic admixture, but also demonstrated a strong imprint of the massive European immigration wave in the 19th and 20th centuries. The high individual ancestral variability observed suggests that each Brazilian has a singular proportion of Amerindian, European and African ancestries in his mosaic genome. In Brazil, one cannot predict the color of persons from their genomic ancestry nor the opposite. Brazilians should be assessed on a personal basis, as 190 million human beings, and not as members of color groups.
Key words: DNA tests; Insertion-deletion polymorphisms; Ancestry; Human diversity; Brazilians
Introduction
Introduction
Brazilians are one of the most heterogeneous populations in the world, the result of five centuries of interethnic crosses between peoples from three continents: Amerindians, Europeans and Africans. Little is known about the number of indigenous people living in the area of what is now Brazil when the Portuguese arrived in 1500 (1), although a figure often cited is that of 2.5 million individuals (2). The Portuguese-Amerindian admixture started soon after the arrival of the first colonizers and later became commonplace, being even encouraged after 1755 as a strategy for population growth and colonial occupation of the country (3).
From the middle of the 16th century, Africans were brought to Brazil to work on sugarcane farms and, later, in the gold and diamond mines and on coffee plantations. Historical records suggest that between circa 1550 and 1850 (when the slave trade was abolished), around four million Africans arrived in Brazil (2).
In reference to the European immigration, it is estimated that about 500,000 Portuguese arrived in the country between 1500 and 1808 (2). From then on, the Brazilian ports were legally opened to all friendly nations.
Significantly, in the approximately 100-year period from 1872 to 1975, Brazil received at least 5.5 million other immigrants from Europe and other parts of the world (2). These were, in decreasing order, 34% Italians, 29% Portuguese, 14% Spanish, 5% Japanese, 4% Germans, 2% Lebanese and Syrians, and 12% others. This phenomenon, which has been denominated the "whitening of Brazil", had complex economic and sociological causes, was tinged with racist ideology, and has been well discussed in the literature (4,5).
It is worthwhile to open a small parenthesis to point out that modern humanity had a single origin in Africa circa 200,000 years ago (reviewed in Ref. 6). Europeans and Asians descend from a relatively small group that migrated out of Africa roughly 60,000 years before the present and thus are genealogically related to Africans, rather than constituting biologically distinct groups (7). Brazil might be seen as representing a "meeting point" for the three major historical geographical components of humanity (Africans, Asians, represented by their Native American descendants, and Europeans) that separated in the first out-of-Africa diaspora.
In such reunion, we can roughly envisage three periods. Initially, the largest population component consisted of the indigenous Amerindians, who thus contributed heavily to the initial formation of Brazilians. The subsequent decrease in the number of Amerindians by the combined effect of guns, germs and steel (8) and the large influx of Africans from the slave trade led to a second phase that lasted until 1850. The third period occurred after 1850 when African immigration stopped and the very prominent entry of Europeans occurred, leading to the "whitening" of Brazil (4,5).
Genetic variation in Brazilians
In the past few years we have been using several different molecular tools to try to characterize the ancestry and formation of the Brazilian people. We will briefly describe these studies, from which we could unravel evidence of genetic admixture at much higher levels than had previously been suspected. Unfortunately, size limitations do not allow us to review the whole field, especially the many important contributions of other research groups.
Uniparental genetic markers in Brazilians
There are several types of genetic markers at the DNA level and they can be classified according to their molecular nature and genomic localization. Because of extensive recombination, autosomal haplotypes are evanescent, constituting excellent individuality markers.
On the other hand, uniparental maternal (mitochondrial DNA, mtDNA) and paternal (non-recombinant region of the Y chromosome, NRY) polymorphisms are excellent stable lineage markers because they are haploid and do not undergo recombination. As such, blocks of genes (haplotypes) transmitted to the next generations remain unaltered in the matrilineages and patrilineages until a mutation supervenes. The new mutations that have occurred and reached high frequencies after the dispersion of modern man from Africa can be specific to certain regions of the globe and can serve as geographical markers.
The mitochondrial DNA and the NRY provide complementary information that can be traced back to several generations and allow the reconstitution of the history of a nation through the migrations of women and men, respectively. Lineage markers can also be employed at the individual level, with the caveat that mtDNA and NRY will only provide information about a single woman or man ancestor of an individual among thousands of ancestors, hence making up a small proportion of the genetic constitution of a person. However, when lineage markers are applied to sets of people, they provide reliable information about the range, composition and proportions of ancestral roots (that we might call the Ancestrome) of the groups.
We initially examined DNA polymorphisms in the non-recombining portion of the Y chromosome to investigate the contribution of distinct patrilineages to the present-day white Brazilian population. Y chromosome polymorphisms were typed in 200 unrelated males from four geographical regions of Brazil and in 93 Portuguese males. In our Brazilian sample, the vast majority of Y chromosomes proved to be of European origin - only 2% of the Y chromosome lineages were from Sub-Saharan Africa (haplogroup E3a*), and none was Amerindian (haplogroup Q3*) (9,10).
There was no significant differentiation among the proportions of Y lineages of the four geographical regions of Brazil. Likewise, there were no significant differences when the haplogroup frequencies in Brazil and Portugal were compared by means of exact tests of population differentiation (11). Nevertheless, by typing with fast evolving NRY markers we later could uncover a higher within-population haplotype diversity in Brazil than in Portugal, explainable by the input of European Y chromosomes of diverse origins (11).
To learn about the maternal counterpart, we analyzed mtDNA, which revealed a different reality. Considering Brazil as a whole, 33, 39, and 28% of matrilineages were of Amerindian, European and African origin, respectively (9,12). As expected, the frequency of different regions reflected their genealogical histories: most matrilineal lineages in the Amazon region were of Amerindian origin, while African ancestrality was preponderant in the Northeast (44%) and European haplogroups were prevalent in the South (66%). These data have since been amply confirmed by numerous other studies. For instance, we recently analyzed the mtDNA haplogroup structure of 242 self-identified white individuals from São Paulo and ascertained 24% Amerindian, 22% African and 54% European matrilineal proportions (Dornelas HG, Bydlowski SP, Pena SDJ, unpublished data).
Next, for further confirmation, we studied mtDNA lineages in 120 black individuals from the city of São Paulo (13). The results, as expected, showed a mirror image of those previously found in white Brazilians: on the one hand, 85% of the lineages originated in Sub-Saharan Africa, 12% were from Amerindians and only 3% were from Europe; on the other, only 48% of the Y chromosome lineages originated from Sub-Saharan Africa (the vast majority belonging to haplogroups E3a7 and E3a*). Studies on black individuals from the cities of Rio de Janeiro and Porto Alegre (14) produced very similar results.
Taken together, these numbers disclose a picture of very strong directional mating between European males and Amerindian and African females, which agrees perfectly with the known history of the peopling of Brazil since 1500. These studies also reveal that the genomes of most Brazilians are mosaic, having mtDNA and NRY of different phylogeographical origins.
Biparental genetic markers and ancestry in Brazilians
In Brazil, notwithstanding relatively large levels of genetic admixture and a myth of "racial democracy", there exists widespread social prejudice that seems to be particularly connected to the physical appearance of an individual (15). Color (in Portuguese, cor) denotes the Brazilian equivalent of the English term race (raça) and is based on a complex phenotypic evaluation that takes into account, besides skin pigmentation, also hair type, nose shape and lip shape (16). The reason why the word color is preferred to race in Brazil is probably because it captures the continuous aspects of phenotypes (16). In contrast with the situation in the United States, there appears to be no racial descent rule operational in Brazil and it is possible for two siblings differing in color to belong to completely diverse racial categories (15).
Based on the criteria of self-classification of the 2000 census of the Instituto Brasileiro de Geografia e Estatística (IBGE), the Brazilian population was then composed of 53.4% Whites, 6.1% Blacks and 38.9% Brown ("pardos" in Portuguese). How do these numbers correlate with genomic ancestry?
Inferences about the European and African ancestral genomic roots. Using a panel of genetic polymorphisms that display large differences in allelic frequencies (>0.40; these polymorphisms are called ancestry-informative markers, or AIMs for short) between Europeans and Africans, Parra et al. (17) showed that, at the population level, it was possible to estimate with great precision the degree of European and African ancestry among North Americans. We decided to ascertain whether this same panel of markers would be able to estimate the degree of African ancestry in Brazilians at the individual level. For this purpose, we selected ten of the best AIMs used in the American study (18).
In order to determine the individual discrimination power of this set of 10 AIMs, we initially genotyped a small sample of individuals from the Northern part of Portugal and from the island of São Tomé, located in the Gulf of Guinea, on the west coast of Africa. These population sources were chosen because they are geographically related to the European and African population groups that participated in the peopling of Brazil. A complete individual discrimination between the European and African genomes was obtained. It was thus clear that the 10-allele set of Parra et al. (17) was highly efficient and provided reliable individual discrimination between European and African genomes.
Our initial Brazilian sample consisted of 173 individuals from a Southeastern rural community, clinically classified according to their color (white, black, or intermediate) with a multivariate evaluation based on skin pigmentation in the medial part of the arm, hair color and texture, and the shape of the nose and lips. When we compared the African genomic ancestry values assessed for these individuals, we observed that the groups had much wider ranges than those of Europeans and Africans and that there was a very significant overlap between them. This indicated that in Brazil there is significant dissociation of color and genomic ancestry, i.e., at the individual level it was not possible to infer the ancestry of a Brazilian from his/her color (18).
To corroborate these findings, we undertook a second investigation based on data from 12 forensic microsatellites that had been used to estimate the personal genomic origin of each of 752 individuals from the city of São Paulo, belonging to different Brazilian color categories (275 Whites, 192 Browns and 285 Blacks) (19). The genotypes permitted the calculation of a personal likelihood-ratio estimator of European or African ancestry. Again, we observed great overlaps among the color categories of Brazilians. This was confirmed quantitatively using a Bayesian analysis of population structure that did not demonstrate significant genetic differentiation between the color groups. These results corroborate and validate our previous conclusions using ancestry-informative markers.
If we consider some peculiarities of Brazilian history and social structure, we can construct a model to explain why color should indeed be a poor predictor of African ancestry (18). Nowadays most Africans have black skin, genetically determined by a very small number of genes that were evolutionarily selected in adaptation to the tropical and subtropical climate. Thus, if we have a social race identification system based primarily on phenotype, as is the case for Brazil, we classify individuals on the basis of the presence of certain alleles in a small number of genes that have an impact on physical appearance, while ignoring the rest of the genome. Assortative mating based on color, which has been shown by demographic studies to occur in Brazil, will produce strong associations among the individual components of color. Indeed, we detected the presence of such positive associations at highly significant levels in a Southeastern Brazilian population (18). On the other hand, we expect that any initial admixture association between color and the AIMs will inevitably decay over time because of genetic admixture. It is easy to see how this combination of social forces could produce a population with distinct color groups and yet with similar levels of African ancestry.
Genomic studies of the Amerindian, European and African ancestral roots. The two studies mentioned above did not take into account the Amerindian contribution to the Brazilian population. To achieve that we needed new polymorphic markers that would be sensitive to all three ancestries.
We screened the database of 2000 human diallelic short indels characterized by Weber et al. (20) and identified 40 polymorphisms that fulfilled the following criteria: widespread chromosomal location in the human genome, increasing amplicon sizes that allowed multiplex PCR amplification and electrophoretic analysis, and allele frequency close to 0.5 in the European population. We used these 40 indel markers to study worldwide human genome variation, namely all the samples in the CEPH-HGDP Diversity Panel (21), composed of 52 populations originating from five geographical regions: Americas, Sub-Saharan Africa, East Asia, Oceania and a cluster composed of Europe, the Middle East and Central Asia (22). We obtained a distance matrix for the 52 populations using the Reynolds genetic measure, which is based on the FST linearized for short divergence times (23). By visualization using Multidimensional Scaling analysis (24), we obtained a very adequate graphic representation that showed five widely separated clusters corresponding to Africa, Oceania, East Asia, Americas and a central European-Middle East-Central Asian group (Figure 1).
Next, we submitted the genotypes of the Amerindians, Europeans and Sub-Saharan Africans of the CEPH panel (the three Brazilian ancestral roots) to the Structure program, version 2.1 (25), a Bayesian software that uses multilocal genotypes to infer the structure of population and group individuals on the basis of their genotypes even without any prior information on the population origin of each sampled individual. The program produced a triangular plot in which the three different populations clustered in different vertices with no overlap (Figure 2). European individuals had on average 94.6% European ancestry, Sub-Saharan Africans had on average 96.5% Sub-Saharan African ancestry and Amerindians had on average 94.8% Amerindian ancestry (Bastos-Rodrigues L, Pimenta JR, Bydlowski S, Pena SDJ, unpublished data).
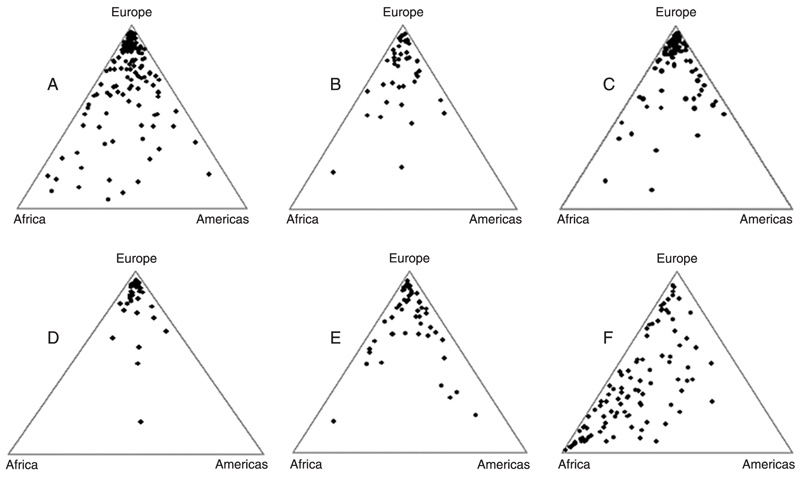
Inferences about the Amerindian, European, and African ancestral genomic roots among Brazilians. As shown in the previous section, Amerindian, African, and European samples of CEPH can be used to define a triangular landscape with ancestry-specific vertices on which we can plot the results of Structure analyses of Brazilian samples. Figure 3 shows triangular plots for Brazilian self-declared White individuals from Minas Gerais in the Southeast Region, North region (States of Amazonas, Acre, Rondônia, and Pará), State of Pernambuco in the Northeast Region, South region (States of Rio Grande do Sul, Santa Catarina, and Paraná), and the city of São Paulo in the Southeast Region. The last graph (F) shows 100 self-declared Black Brazilian individuals from the city of São Paulo.
We can observe from the figure that most of the white individuals of all the regions examined clustered in the "European" vertex of the triangle plot (Figure 3A-E), although a proportion of them were scattered throughout the triangle area. As can be checked in Table 1, all average European contributions to white individuals were above 0.700, with a maximum of 0.819 in Southern Brazil (a region of heavy European immigration) and a minimum of 0.709 in Minas Gerais. When we compared the regions pairwise using a Monte Carlo resampling strategy (26), however, we could not find statistical significance. We then pooled all the samples of Brazilian Whites and compared it with the sample of Europeans from the CEPH panel, now finding a significant difference (P < 0.0001).
Black individuals from the city of São Paulo showed very high individual variation in their biogeographical ancestry, as indicated by a large spread of data points in the triangular graph (Figure 3F). They had an average degree of African ancestry slightly below 50% (Table 1), which was significantly different from white individuals of the same region and also from individuals from Sub-Saharan Africa.
An interesting observation is that the extent of Amerindian ancestry is relatively low (range 0.092-0.147) and not statistically different among White individuals from different geographical regions and also between White and Black individuals from São Paulo.
Conclusions
Many investigators have used historical, sociological and anthropological methodology to analyze the origins of Brazilians: Paulo Prado in Retrato do Brasil (published in 1927), Sérgio Buarque de Holanda in Raízes do Brasil (published in 1933), Gilberto Freyre in Casa Grande e Senzala (published in 1933), and Darcy Ribeiro in O Povo Brasileiro (27). We have used new molecular genetics tools for the same purpose.
The data presented in this review demonstrate that currently available DNA tests can provide an important molecular confirmation of the proposals of the authors mentioned above and are also capable of providing new valuable insights into the process of genetic formation and structure of the Brazilian people.
Studies with uniparental markers in both white and black Brazilians demonstrate strong directional mating between European males and Amerindian and African females, which agrees with the known history of the peopling of Brazil since 1500. These data reveal that the genomes of most Brazilians are mosaic, having mtDNA and NRY of different phylogeographical origins.
Studies with autosomal biparental markers reveal very elevated levels of genetic admixture between the three ancestral roots. However, it is also evident that there was an important population effect of the program of "whitening" of Brazil promoted through the immigration of circa six million Europeans in the roughly 100-year period after 1872. This manifests itself both in a predominant (>70%) European genomic ancestry in Brazilian Whites regardless of geographical region and in a high average European genomic ancestry (37.1%) in Brazilian Blacks.
The correlation between color and genomic ancestry is imperfect: at the individual level one cannot safely predict the skin color of a person from his/her level of European, African and Amerindian ancestry nor the opposite. Regardless of their skin color, the overwhelming majority of Brazilians have a high degree of European ancestry. Also, regardless of their skin color, the overwhelming majority of Brazilians have a significant degree of African ancestry. Finally, most Brazilians have a significant and very uniform degree of Amerindian ancestry!
The high ancestral variability observed in Whites and Blacks suggests that each Brazilian has a singular and quite individual proportion of European, African and Amerindian ancestry in his/her mosaic genomes. Thus, the only possible basis to deal with genetic variation in Brazilians is not by considering them as members of color groups, but on a person-by-person basis, as 190 million human beings,with singular genome and life histories.
To test the discrimination power of our 40-indel set, we obtained a distance matrix of the 52 populations of the CEPH-HGDP international sample set (21) using the Reynolds genetic measure, which is based on the FST linearized for short divergence times (24). From the matrix we undertook a Multidimensional Scaling analysis (24) using the Statistica program. It is immediately apparent that the resolution power was excellent and that the individuals from the 52 populations aggregate into five widely separated clusters that correspond to Africa, Oceania, East Asia, Americas, and a central Europe-Middle East-Central Asia group (modified from Ref. 22).
Triangular plot produced by the Structure program, version 2.1 (25), on analysis of the genotypes of the European, Amerindian and Sub-Saharan African individuals of the CEPH-HGDP panel (the three Brazilian ancestral roots). Structure uses multilocal genotypes to group individuals on the basis of their genotypes. The run consisted of 100,000 burn-in steps, followed by 2.5 x 105 Markov Chain Monte Carlo iterations, without any prior information on the population origin of each individual sampled. We used the "admixture" model and assumed the allele frequencies of different populations to be correlated.
Triangular graphs of the genomic proportions of Amerindian, European and African ancestry of Brazilian individuals drawn using the Tri-Plot program (28). We used our set of 40 insertion-deletion polymorphisms (22) and the Structure program to study 272 Brazilians self-defined as White (11,18). A, 142 individuals from the State of Minas Gerais in the Southeast; B, 45 individuals from the North (States of Amazonas, Acre, Rondônia, and Pará); D, 36 individuals from the South (States of Rio Grande do Sul, Santa Catarina and Paraná); E, 49 individuals from the Northeast (State of Pernambuco). We also studied 88 White (C) and 100 Black (F) men from the city of São Paulo, Brazil, randomly drawn from a larger sample described in a previous publication (19).
Average values of the genomic proportions of Amerindian, European and African ancestry of individuals from the CEPH-HGDP panel (21) and from the Brazilian population (see description in the legend to Figure 3).
 Address for correspondence: S.D.J. Pena, GENE - Núcleo de Genética Médica, Av. Afonso Pena, 3111/9, 30130-909 Belo Horizonte, MG, Brasil. Fax: +55-31-3227-3792. E-mail: spena@gene.com.br
Address for correspondence: S.D.J. Pena, GENE - Núcleo de Genética Médica, Av. Afonso Pena, 3111/9, 30130-909 Belo Horizonte, MG, Brasil. Fax: +55-31-3227-3792. E-mail: spena@gene.com.br
Research supported by CNPq and a Millenium Institute Project of the Ministério de Ciência e Tecnologia of Brazil on Genomic Ancestry (coordinator: F.M. Salzano). Received July 23, 2009. Accepted August 18, 2009. Available Online September 11, 2009.
The Brazilian Journal of Medical and Biological Research is partially financed by
References
- 1. Vainfas R. História indígena: 500 anos de despovoamento. In: IBGE (Editor), Brasil: 500 anos de povoamento. Rio de Janeiro: Instituto Brasileiro de Geografia Estatística - IBGE; 2000. p 37-59.
- 2. IBGE. Brasil - 500 anos de povoamento. Rio de Janeiro: Instituto Brasileiro de Geografia Estatística - IBGE; 2000.
- 3. Salzano FM, Freire-Maia N. Problems in human biology: a study of Brazilian populations. Detroit: Wayne State University Press; 1970.
- 4. Seyferth G. A antropologia e a teoria do branqueamento de raça no Brasil: a tese de João Batista de Lacerda. Rev Mus Paulista 1985; 30: 81-98.
- 5. Santos AS. Historical roots of the "Whitening" of Brazil. Lat Am Perspect 2002; 28: 61-82.
- 6. Pena SDJ. The evolution and structure of human genetic diversity. In: Suarez-Kurtz G (Editor), Pharmacogenomics in admixed populations. Austin: Landes Bioscience; 2009. p 1-11.
- 7. Hunley KL, Healy ME, Long JC. The global pattern of gene identity variation reveals a history of long-range migrations, bottlenecks, and local mate exchange: implications for biological race. Am J Phys Anthropol 2009; 139: 35-46.
- 8. Diamond J. Guns, germs, and steel: the fates of human societies. New York: WW Norton & Co.; 1997.
- 9. Pena SDJ, Carvalho-Silva DR, Alves-Silva J, Prado VF, Santos FR. Retrato molecular do Brasil. Ciência Hoje 2000; 27: 16-25.
- 10. Carvalho-Silva DR, Santos FR, Rocha J, Pena SD. The phylogeography of Brazilian Y-chromosome lineages. Am J Hum Genet 2001; 68: 281-286.
- 11. Carvalho-Silva DR, Tarazona-Santos E, Rocha J, Pena SD, Santos FR. Y chromosome diversity in Brazilians: switching perspectives from slow to fast evolving markers. Genetica 2006; 126: 251-260.
- 12. Alves-Silva J, da Silva Santos M, Guimaraes PE, Ferreira AC, Bandelt HJ, Pena SD, et al. The ancestry of Brazilian mtDNA lineages. Am J Hum Genet 2000; 67: 444-461.
- 13. Goncalves VF, Carvalho CM, Bortolini MC, Bydlowski SP, Pena SD. The phylogeography of African Brazilians. Hum Hered 2008; 65: 23-32.
- 14. Hunemeier T, Carvalho C, Marrero AR, Salzano FM, Junho Pena SD, Bortolini MC. Niger-Congo speaking populations and the formation of the Brazilian gene pool: mtDNA and Y-chromosome data. Am J Phys Anthropol 2007; 133: 854-867.
- 15. Harris M, Kotak C. The structural significance of Brazilian categories. Sociologia 1963; 25: 203-208.
- 16. Telles EE. Racial ambiguity among the Brazilian population. Ethn Racial Stud 2002; 25: 415-441.
- 17. Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet 1998; 63: 1839-1851.
- 18. Parra FC, Amado RC, Lambertucci JR, Rocha J, Antunes CM, Pena SD. Color and genomic ancestry in Brazilians. Proc Natl Acad Sci U S A 2003; 100: 177-182.
- 19. Pimenta JR, Zuccherato LW, Debes AA, Maselli L, Soares RP, Moura-Neto RS, et al. Color and genomic ancestry in Brazilians: a study with forensic microsatellites. Hum Hered 2006; 62: 190-195.
- 20. Weber JL, David D, Heil J, Fan Y, Zhao C, Marth G. Human diallelic insertion/deletion polymorphisms. Am J Hum Genet 2002; 71: 854-862.
- 21. Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, et al. A human genome diversity cell line panel. Science 2002; 296: 261-262.
- 22. Bastos-Rodrigues L, Pimenta JR, Pena SD. The genetic structure of human populations studied through short insertion-deletion polymorphisms. Ann Hum Genet 2006; 70: 658-665.
- 23. Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 1983; 105: 767-779.
- 24. Kruskal JB, Wish M. Multidimensional Scaling. New York: SAGE Publications; 1978.
- 25. Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000; 155: 945-959.
- 26. Manly BFJ. Randomization, bootstrap and Monte Carlo methods in biology. New York: Chapman & Hall; 1997.
- 27. Ribeiro D. O povo brasileiro: a formação e o sentido do Brasil. São Paulo: Companhia das Letras; 1995.
- 28. Graham DJ, Midgley NG. Graphical representation of particle shape using triangular diagrams: an Excel spreadsheet method. Earth Surf Proc Landf 2000; 25: 1473-1477.
Publication Dates
-
Publication in this collection
11 Sept 2009 -
Date of issue
Oct 2009
History
-
Accepted
18 Aug 2009 -
Received
23 July 2009

 DNA tests probe the genomic ancestry of Brazilians
DNA tests probe the genomic ancestry of Brazilians