Abstract
Novel coronavirus (nCoV) namely “SARS-CoV-2” is being found responsible for current PANDEMIC commenced from Wuhan (China) since December 2019 and has been described with epidemiological linkage to China in about 221 countries and territories until now. In this study we have characterized the genetic lineage of SARS-CoV-2 and report the recombination within the genus and subgenus of coronaviruses. Phylogenetic relationship of thirty nine coronaviruses belonging to its four genera and five subgenera was analyzed by using the Neighbor-joining method using MEGA 6.0. Phylogenetic trees of full length genome, various proteins (spike, envelope, membrane and nucleocapsid) nucleotide sequences were constructed separately. Putative recombination was probed via RDP4. Our analysis describes that the “SARS-CoV-2” although shows great similarity to Bat-SARS-CoVs sequences through whole genome (giving sequence similarity 89%), exhibits conflicting grouping with the Bat-SARS-like coronavirus sequences (MG772933 and MG772934). Furthermore, seven recombination events were observed in SARS-CoV-2 (NC_045512) by RDP4. But not a single recombination event fulfills the high level of certainty. Recombination mostly housed in spike protein genes than rest of the genome indicating breakpoint cluster arises beyond the 95% and 99% breakpoint density intervals. Genetic similarity levels observed among “SARS-CoV-2” and Bat-SARS-CoVs advocated that the latter did not exhibit the specific variant that cause outbreak in humans, proposing a suggestion that “SARS-CoV-2” has originated possibly from bats. These genomic features and their probable association with virus characteristics along with virulence in humans require further consideration.
Keywords: COVID-19; Novel coronavirus (nCoV); Bat-SARS-CoVs; phylogenetic relationship; Recombination detection program-4
Resumo
O novo coronavírus (nCoV), nomeadamente “SARS-CoV-2”, foi considerado responsável pela pandemia atual iniciada em Wuhan (China) desde dezembro de 2019 e foi descrito com ligação epidemiológica à China em cerca de 221 países e territórios até agora. Neste estudo, caracterizamos a linhagem genética do SARS-CoV-2 e relatamos a recombinação dentro do gênero e subgênero dos coronavírus. A relação filogenética de 39 coronavírus pertencentes a seus quatro gêneros e cinco subgêneros foi analisada usando o método de Neighbour-joining usando MEGA 6.0. Árvores filogenéticas do genoma de comprimento total, várias proteínas (espícula, envelope, membrana e nucleocapsídeo), sequências de nucleotídeos foram construídas separadamente. A recombinação putativa foi testada via RDP4. Nossa análise descreve que o “SARS-CoV-2”, embora mostre grande semelhança com as sequências de Bat-SARS-CoVs em todo o genoma (dando semelhança de sequência de 89%), exibe agrupamento conflitante com as sequências de coronavírus do tipo Bat-SARS (MG772933 e MG772934) Além disso, sete eventos de recombinação foram observados em SARS-CoV-2 (NC045512) por RDP4. Mas nem um único evento de recombinação preenche o alto nível de certeza. A recombinação está alojada mais em genes de proteína de pico, principalmente, do que no resto do genoma, indicando que o cluster de ponto de interrupção surge além dos intervalos de densidade de ponto de interrupção de 95% e 99%. Os níveis de similaridade genética observados entre “SARS-CoV-2” e Bat-SARS-CoVs defendem que o último não exibe a variante específica que causa surto em humanos, sugerindo que “SARS-CoV-2” tenha se originado possivelmente de morcegos. Essas características genômicas e sua provável associação com as características do vírus, juntamente com a virulência em humanos, requerem uma consideração mais aprofundada.
Palavras-chave: Covid-19; Novo coronavírus (nCoV); Bat-SARS-CoVs; relação filogenética; Programa de detecção de recombinação-4
1. Introduction
World Health Organization (WHO) defines, continuous emergences of viral ailments possess a serious problem to the public health. During preceding two decades, many particular viral epidemics like severe acute respiratory syndrome coronavirus (SARS-CoV) was noted (2002-2003) similarly, H1N1 influenza (2009). However, Middle East Respiratory Syndrome Coronavirus (MERS-CoV) was latest and firstly recognized in Saudi Arabia (2012). These epidemics like SARS-CoV triggered an epidemic in China affecting many other countries with about 8,000 cases similarly MERS-CoV has about 2,500 cases while about 8,00 deaths in both (Cascella et al., 2020).
During whole period which extends till today, an epidemic characterized by inexplicable low respiratory infections was identified in a largest cosmopolitan area in Wuhan (Hubei province), was first time reported on December 31, 2019, to the Country Office of WHO in China. However, some published literature showed the commencement of symptoms exhibiting cases back to the beginning of December, 2019. The causative agent was unidentified so; initial cases were called as “pneumonia of unknown etiology.” The Chinese Center for Disease Control and Prevention (CDC) and few local CDCs established a complete program for outbreak investigation. The causative agent of the etiology of this infection is a “novel” virus member of coronavirus (CoV) family. Firstly, it was called 2019-nCoV, latter on International Committee on Taxonomy of Viruses (ICTV) termed it as “SARS-CoV-2” virus due to its great similarity with that virus caused SARS outbreak (SARS-CoVs) (Gorbalenya et al., 2020).
Dr. Tedros Adhanom Ghebreyesus; Director-General of WHO, declared on 11th February, 2020 that current illness was called as “COVID-19,” an acronym of “coronavirus disease 2019” caused by novel CoV, a very contagious and has spread quickly around the world. Another turning point appeared on 26th February, 2020, as another first case which was not imported from China, noted in the United States. On 30th January 2020, in a meeting, per the International Health Regulations (IHR), WHO announced this outbreak a “Public Health Emergency of International Concern (PHEIC)” due to its extensive spreading and transmission from human-to-human. On March 11, COVID-19 cases increased many folds involving numerous countries and many deaths, WHO proclaimed the COVID-19 as a “PANDEMIC” (Takian et al., 2020; Cascella et al., 2020).
The CoVs are from a huge family of single-stranded RNA viruses (+ssRNA) being segregated from various species of animals (Perlman and Netland 2009). CoVs appeared crown-like derived from coronam a Latin term means crown, as its envelope contain spike of glycoproteins and are positive-stranded RNA viruses. The subfamily Orthocoronavirinae of the Coronaviridae; order Nidovirales, categorizes into four genera: 1 (Alphacoronavirus: alphaCoV), 2 (Betacoronavirus: betaCoV), 3 (Deltacoronavirus: deltaCoV) and 4 (Gammacoronavirus: gammaCoV). Probable gene sources of first two genera are from bats and rodents and last two from avian species. Moreover, genus betaCoV splits into five further sub-genera/lineages (Chan et al., 2013).
Its genome has ~30,000 nucleotides and has elliptic/round and frequently pleomorphic form with diameter of about 60-140 nm with 5’ Cap and 3’ poly (A) tail. At least six ORFs constitute the genome and subgenomes of a representative CoV. The first ORFs (ORF1a/b), which encode 16 non-structural proteins (nsp1‐16), except Gammacoronavirus that lacks nsp1 represents 2/3 of the whole genome length. There is a (−1) frameshift among ORF1a which lead to the production of pp1a while, ORF1b responsible for the production of pp1ab polypeptide. Latter on these polypeptides are being processed via virally encoded one or two papain‐like protease, chymotrypsin‐like protease (3CLpro) and or main protease (Mpro) into 16 nsps (Masters 2006; Ziebuhr et al. 2000) as shown in Figure 1 and 2. Remaining ORFs located on the 1/3 of the genome nearby the 3′terminus encodes for no less than four main structural proteins {spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins} (as illustrated in Figure 2). All accessory and structural proteins are translated via sgRNAs of CoVs (Hussain et al., 2005).
Diagramtic illustration of position and structure of four structural proteins (spike protein, envelope protein, membrane protein and nucleocapsid protein) of SARS-CoV-2.
The indicated viruses are potent to overcome species barriers along with causing many illnesses in human varying from the “common cold” to very cruel diseases like MERS and SARS, for unknown reasons (Singhal, 2020; Assiri et al., 2013). Most likely, these viruses have originated from bats to alternate mammalian hosts, as SARS-CoV by Himalayan palm civet, and MERS-CoV via dromedary camel, before hopping to humans (Banerjee et al. 2019). However, dynamics of “SARS-CoV-2” are at presented not well known, there are assumptions about its animal origin. Though, its origins are not clear, but genomic analyses advocate that SARS-CoV-2 has possibly evolved from a strain present in bats. So, the mutation in the original strain may have directly activated virulence towards humans. High ability of this virus to become a global pandemic possesses a serious public health risk, as this virus spreads faster than its two ancestors i.e., SARS-CoV and MERS-CoV (Singhal, 2020).
Treatments vary for suspected (needed to be isolated at first glance) and confirmed (required to be shifted to the hospitals) patients. The basic necessity is to sustain hydration, nutrition and controlling fever and cough (Jin et al., 2020; Singhal, 2020; Chen et al., 2020). Considering the need, oxygen therapy, as nasal catheter, High Flow Nasal Oxygen Therapy (HFNO), Non-Invasive Ventilation (NIV), mask oxygen or Extracorporeal Membrane Oxygenation should be administered (ECMO; Jin et al., 2020). Previously, no particular vaccine or antiviral cure for COVID-19 is recommended (Lu et al., 2020). However, use of antibiotics and antivirals must be avoided in confirmed cases (Jin et al., 2020; Singhal, 2020, Chen et al., 2020). Antibiotics and antifungals are required if co-infections occur (Russell et al., 2020; Zhao et al., 2020). Various antiviral drugs as oseltamivir, ganciclovir and lopinavirritonavir (Xu et al., 2020), Lopinavir /Ritonavir, Neuraminidase inhibitors, Nucleoside analogues, abidol, peptide (EK1), RNA synthesis inhibitors (TDF, 3TC), anti-inflammatory agents like hormones and traditional Chinese (Lianhuaqingwen Capsule and ShuFengJieDu) medicines, might be used (Lu et al., 2020; Jin et al., 2020). Remdesivir as well as chloroquine (Wang et al., 2020), arbidol an antiviral drug present in and China and Russia, interferons, intravenous immunoglobulin, and plasma of COVID-19 recovered patients, can be proposed for therapy after more evidences of efficacy and safety (Jin et al., 2020; Zhang and Liu, 2020). Currently, different vaccines viz., moderna, Pfizer-BioNTEC, Johnson & Johnson/Janssen (sinovac) Sinopharm/Beijing, Sputnik V and Oxford/AstraZeneca have been administered in many countries of the world (Our World in Data, 2021).
2. Materials and Methods
2.1. Retrieving and acquisition of sequence data
Sequence analysis of “SARS-CoV-2” whole-genome (Genbank accession no. NC_045512) belongs to betacoronavirus and its full genome sequence analysis shown that it is deviating from MERS-CoV and SARS-CoV (Zhu et al., 2020) as shown in Figure 3. The “SARS-CoV-2” as well as Bat-SARS-CoVs exhibits a distinctive lineage inside the subgenus of the sarbecovirus (Zhu et al., 2020). We were aiming to characterize the genomic relationships of full length genome and four open reading frames {spike protein (S), envelope protein (E), membrane protein (M) and nucleocapsid protein (N)} of “SARS-CoV-2” and exploration of putative recombination. Thirty nine full viral sequences that belong to four genera of CoVs viz. alpha-CoVs, beta-CoVs, gamma-CoVs, delta-CoVs and five subgenus of beta CoVs were downloaded from NCBI nucleotide sequence database (NCBI, 2021). Various accession number, organism, host, country and collection year of all thirty nine CoVs sequences (mentioned in Supplementary Material Table 1). These sequences were selected from the research articles (Zhou et al., 2020; Paraskevis et al., 2020; Wang et al., 2018). Each genome was segregated into S, E, M and N for phylogenetic, recombination and mutational analyses.
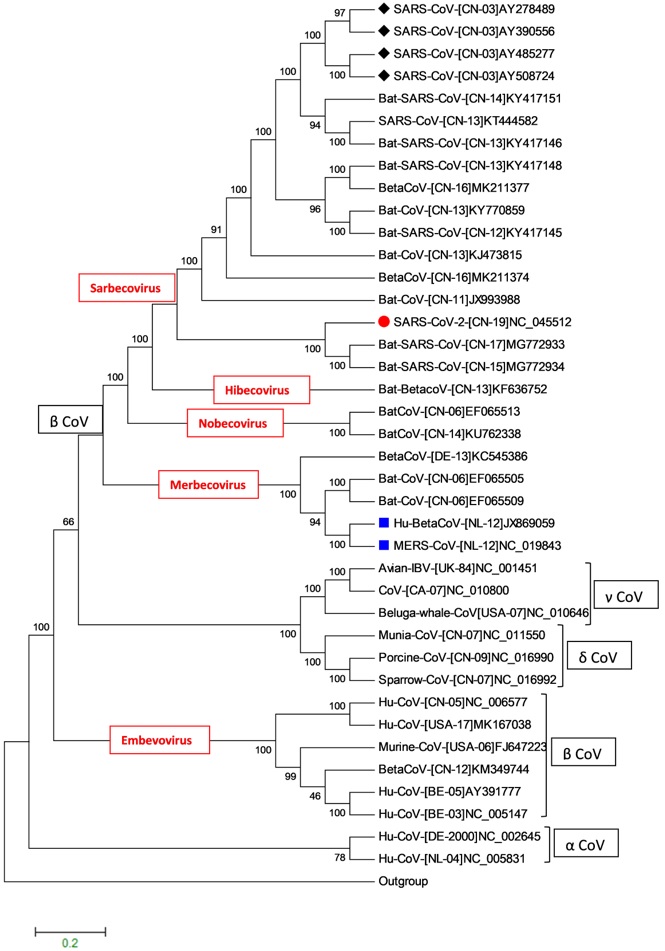
Phylogenetic tree generated based on an alignment of the coronaviruses full genomic sequences available in the GenBank databases. ● indicated the SARS-CoV-2, ▄ indicated the MERS-CoV, ♦ indicated the SARS-CoV.
2.2. Phylogenetic tree construction using MEGA 6.0.
For this, sequences were first aligned via CLUSTAL W method of MEGA6 (Tamura et al., 2013) for full length as well as S, E, M and N sequences phylogenetic tree construction, separately. Neighbor-Joining method proposed by Saitou and Nei (1987) was followed for evolutionary history. The optimal tree for full length, S, E, M and N with sum of branch lengths = 7.97034047, 9.34476698, 8.30616966, 8.67787963, 8.54006468 respectively (as shown in Supplementary Material Figure 1 {(a), (b), (c), (d)}. The % age of replicate trees in which the related taxa grouped altogether in the bootstrap test (1000 replicates), shown following branches (Felsenstein, 1985). The tree was drawn to scale, along branch lengths in similar units as those for the evolutionary distances as to deduce the phylogenetic tree. Jukes and Cantor (1969) method was followed for evolutionary distances computation and are in the units of the base substitutions number per site. The rate variation, between sites was modelled via gamma distribution (shape parameter = 5). For analysis 40 nucleotide sequences were involved. Codon locations included were 1st+2nd+3rd+Noncoding. Data missing and gaps containing positions were eradicated. A total of 20914, 1868, 76, 600 and 774 positions in the full length, S, E, M and N final data set were observed. All evolutionary analyses were shown via MEGA6 (Tamura et al., 2013).
2.3. Recombination detection by RDP-4
Recombination Detection Program (RDP4) proposed by Martin et al. (2015) was used for putative recombination by distinguishing probable parents and recombination regions in coronaviruses. Via MEGA6 programming, herein sequences were adjusted then swapped to the RDP-4 to determine recombination. For the evaluation of recombination events default estimation of X-over based on robotized RDP (R), BOOTSCAN (B), GENECONV (G), MaxChi (M), SiScan (S), Chimaera (C), 3SEQ (T), LARD (L) and PhylPro (P). However, a cut off estimation (0.05) was exploited as p-value. Additionally, these recombination events were grasped via phylogenetic investigation of distinct genes alongside nucleotide arrangement by MegAlign (Lasergene, DNA-STAR). Intended for validity, recombinants were further avowed by in excess of one strategy. Additionally, sequences of amino acids of S, E, M and N (GenBank accession no. NC_045512) were retrieved from NCBI to utilized for three-dimensional-modeling of proteins via Phyre2 (Kelley et al., 2015) are presented in Figure 2.
3. Results and Discussion
Phylogenetic analysis exhibited that “SARS-CoV-2” belongs to genus “βCoV” and subgenus “sarbecovirus” along with other SARS and SARS-CoV viruses isolated from human, bat and other animals. Phylogenetic trees of full length, S, E, M and N nucleotide sequences have been constructed separately and thirty nine sequences have been arranged according to their four genera (αCoV, βCoV, δCoV and γCoV) and five subgenus level viz., sarbecovirus, merbecovirus, hibecovirus, embevovirus and nobecovirus. It is depicted from Figure 3 that new coronavirus “SARS-CoV-2” (NC_045512) has close lineage to Bat-SARS-CoVs (MG772933 and MG772934; isolated from the bats from China) than to other human infecting CoVs, including SARS-CoVs and it is not a mosaic (Zhou et al., 2020). Though not only the full genome of Bat-SARS-CoVs remains the closest to SARS-CoV-2 but their S, E, M and N nucleotide sequences also match with each other.
Although it is evident from the BLASTn results that full length, S, E, M and N nucleotide sequences of novel coronavirus and Bat-SARS-CoVs share 89%, 83.68%, 98.68%, 93.92% and 91.28% per cent identity respectively. The spike protein of SARS-CoV-2 has the biggest variation among others, thus revealed the host selectivity. Since, the proteins M and E are involved in virus assembly, whereas the spike protein (S) mediates virus entry into host cells and is a critical determinant of viral host range (Li, 2016). There is a prospective that “SARS-CoV-2” has originated probably from bats CoVs but the levels of their genetic similarity proposes that “SARS-CoV-2” is not an exact variant of old Bat-SARS-CoVs to cause the recent pandemic. Moreover, they showed less than 90% sequence similarity so that quite a long branch in phylogenetic tree is present between them indicating that Bat-SARS-CoVs are not the direct ancestors of SARS-CoV-2. Importantly, we cannot overlook that bats are the natural reservoirs for SARS-CoV and MERS and humans are terminal host whilst masked palm civet and Arabian camels are intermediate hosts of SARS-CoV and MERS respectively. But in case of nCoVs it is assumed that it is hosted by bats and transmitted to human via unknown animal(s) presently (Lu et al., 2020).
Analyses via codon usage can sort out the origin of proteins with deep ancestry and inadequate phylogenetic signals designed de novo. Spike protein of novel coronavirus has close relationship with bat coronaviruses sequences thus showing uniform ancestry (Supplementary Material Figure 1a). Spike (S) protein of novel coronavirus could be result from the recombination of unsampled coronavirus yet unknown (Ji et al., 2020). Moreover, phylogenetic discordance in deep relationships of coronaviruses is common and can be explained either by early recombination event or changed evolutionary rates in different lineages, or a combination of both (Magiorkinis et al., 2004).
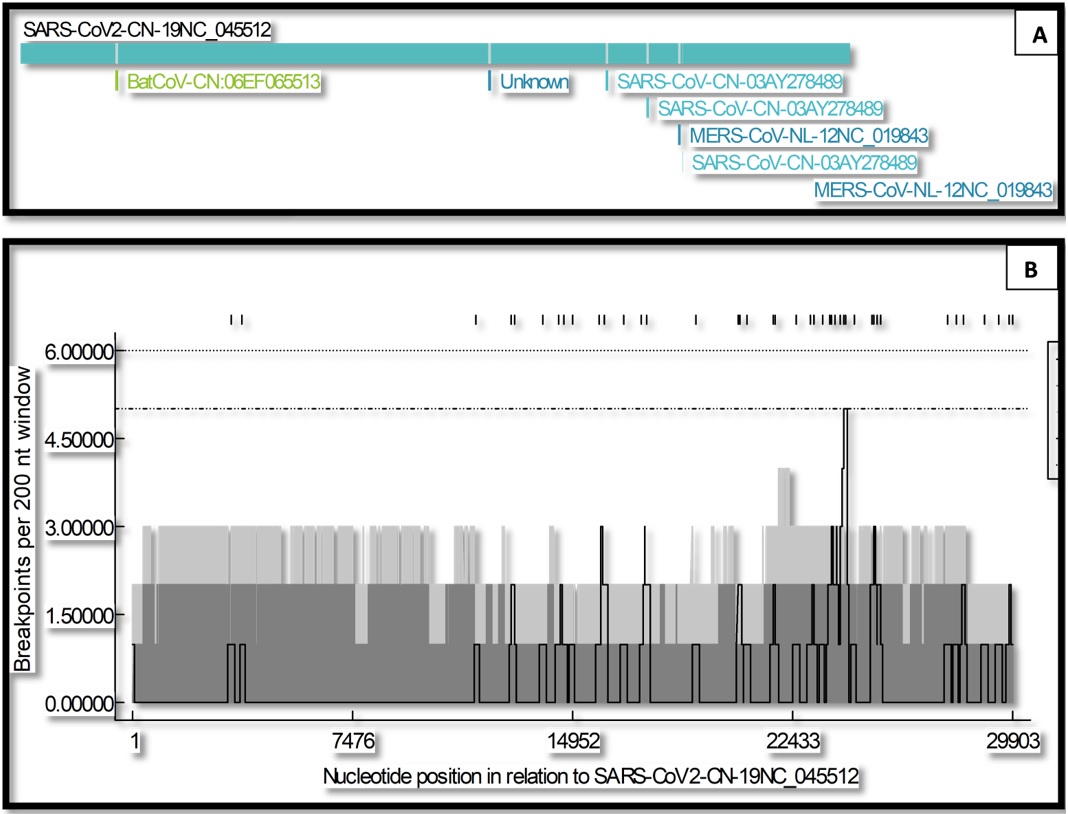
Recombination analysis of ten different coronavirus sequences were characterized using RDP-4. Seven recombination events were observed in SARS-CoV-2 (NC_045512). At least five methods out of nine and recombination score more than 0.50 is required for authenticity. But in this analysis depicted in Table 1, no recombination fulfils the requirement. Moreover, recombination breakpoint distribution maps created between size and distribution of recombination events were noticed in selected coronavirus sequences as described by the Heath et al (2006) and Varsani et al (2008). Many recombination events occurred in spike protein genes else than rest of the genome indicating breakpoint cluster arise more than the 95% and 99% breakpoint density intervals (grey areas) as depicted in Figure 4. Recombination events are multifaceted and frequent in Bat-CoVs than SARS-CoV-2. Henceforth not responsible for nCoVs emergence and COVID-19 outbreak (Lu et al., 2020).
Details of seven recombination events detected in SARS-CoV-2 using RDP-4. Major and minor parents are inferred based on genetic fragments they denoted to the recombinant, with the major parent donating the larger fragment and the minor parent the smaller fragment. Methods used to detect recombination are as follows RDP (R), GENCONV (G), BOOTSCAN (B), MAXCHI (M), CHIMEARA (C), SISCAN (S), PHYLPRO (P), LARD (L) and 3SEQ (T). The method with the most significant associated p-value is indicated in bold for each event.
(A) Recombination analysis of SARS-CoV-2 with other coronaviruses. Seven different recombination events have been identified; (B) Conserved recombination breakpoint distributions in ten selected sequences of coronaviruses. Solid black line represents the recombination breakpoint distribution plot. Y-axis mentioned the breakpoints/200 nucleotide window. Dark filled region represents 95% and light filled region 99% confidence intervals of the ‘local’ recombination breakpoint cold and hot spot test.
Hence, our study nullifies the hypothesis of “SARS-CoV-2” emergence a consequence of recent recombination event. SARS-CoV-2 genome has no close genetic relationship within the subgenus sarbecovirus members, thus provide new lineage. Next-generation sequencing and bioinformatics are changing the way we can respond to pandemic, fast tracking the identification of pathogens, enlightening our understanding of disease incidence and transmission and endorsing data sharing (Armstrong et al., 2019). Moreover, high throughput, unbiased sequencing is a potential technique in the identification of novel pathogens (Palacios et al., 2008).
4. Conclusion
In a nutshell, we have highlighted a detailed genomic structure of SARS-CoV-2 that cause current serious pandemic, coupled with its phylogenetic and recombination analysis with other CoVs belonging to four genera and five subgenera. All the more by and large, the COVID-19 connected to 2019-nCoV features the concealed virus reservoir in wild creature(s) and their capability to once in a while overflow into human populaces. To bring this pandemic to an end, a large share of the world needs to be immune to the virus by vaccination. Now the challenge is to make these vaccines available to people around the world. It will be key that people in all countries not just in rich countries receive the required protection.
Supplementary Material
Supplementary material accompanies this paper.
Supplementary Figure 1. Phylogenetic tree generated based on an alignment of the coronaviruses (a) Spike protein, (b) Envelope protein, (c) membrane protein and (d) Nucleocapsid protein sequences available in the GenBank databases.
Supplementary Table 1. Illustration of Accession number, organism, host, country and collection year of all thirty nine CoVs sequences.
This material is available as part of the online article from http://www.scielo.br/bjb
Acknowledgements
Authors are thankful to Prof. Dr. Farah Khan, Chairperson, department of Botany, Lahore, College for Women University, Lahore, Pakistan for motivating us to write this manuscript.
References
-
ARMSTRONG, G.L., MACCANNELL, D.R., TAYLOR, J., CARLETON, H.A., NEUHAUS, E.B., BRADBURY, R.S., POSEY, J.E. and GWINN, M., 2019. Pathogen genomics in Public Health. The New England Journal of Medicine, vol. 381, no. 26, pp. 2569-2580. http://dx.doi.org/10.1056/NEJMsr1813907 PMid:31881145.
» http://dx.doi.org/10.1056/NEJMsr1813907 -
ASSIRI, A., MCGEER, A., PERL, T.M., PRICE, C.S., AL RABEEAH, A.A., CUMMINGS, D.A., ALABDULLATIF, Z.N., ASSAD, M., ALMULHIM, A., MAKHDOOM, H., MADANI, H., ALHAKEEM, R., AL-TAWFIQ, J.A., COTTEN, M., WATSON, S.J., KELLAM, P., ZUMLA, A.I., MEMISH, Z.A., and KSA MERS-CoV INVESTIGATION TEAM, 2013. Hospital outbreak of Middle East respiratory syndrome coronavirus. The New England Journal of Medicine, vol. 369, no. 5, pp. 407-416. http://dx.doi.org/10.1056/NEJMoa1306742 PMid:23782161.
» http://dx.doi.org/10.1056/NEJMoa1306742 -
BANERJEE, A., KULCSAR, K., MISRA, V., FRIEMAN, M. and MOSSMAN, K., 2019. Bats and coronaviruses. Viruses, vol. 11, no. 1, pp. 41. http://dx.doi.org/10.3390/v11010041 PMid:30634396.
» http://dx.doi.org/10.3390/v11010041 - CASCELLA, M., RAJNIK, M., CUOMO, A., DULEBOHN, S.C. and DI NAPOLI, R., 2020. Features, evaluation and treatment coronavirus (COVID-19) Treasure Island (FL): StatPearls Publishing.
-
CHAN, J.F., TO, K.K., TSE, H., JIN, D.Y. and YUEN, K.Y., 2013. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends in Microbiology, vol. 21, no. 10, pp. 544-555. http://dx.doi.org/10.1016/j.tim.2013.05.005 PMid:23770275.
» http://dx.doi.org/10.1016/j.tim.2013.05.005 -
CHEN, Y., LIU, Q. and GUO, D., 2020. Emerging coronaviruses: genome structure, replication, and pathogenesis. Journal of Medical Virology, vol. 92, no. 4, pp. 418-423. http://dx.doi.org/10.1002/jmv.25681 PMid:31967327.
» http://dx.doi.org/10.1002/jmv.25681 -
CORMAN, V.M., KALLIES, R., PHILIPPS, H., GÖPNER, G., MÜLLER, M.A., ECKERLE, I., BRÜNINK, S., DROSTEN, C. and DREXLER, J.F., 2014. Characterization of a novel betacoronavirus related to Middle East respiratory syndrome coronavirus in European hedgehogs. Journal of Virology, vol. 88, no. 1, pp. 717-724. http://dx.doi.org/10.1128/JVI.01600-13 PMid:24131722.
» http://dx.doi.org/10.1128/JVI.01600-13 -
CORONAVIRIDAE STUDY GROUP OF THE INTERNATIONAL COMMITTEE ON TAXONOMY OF VIRUSES, GORBALENYA, A.E., BAKER, S.C., BARIC, R.S., DE GROOT, R.J., DROSTEN, C., GULYAEVA, A.A., HAAGMANS, B.L., LAUBER, C., LEONTOVICH, A.M., NEUMAN, B.W., PENZAR, D., PERLMAN, S., POON, L.L.M., SAMBORSKIY, D.V., SIDOROV, I.A., SOLA, I. and ZIEBUHR, J., 2020. The species severe acute respiratory syndrome related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology, vol. 5, no. 4, pp. 536-544. http://dx.doi.org/10.1038/s41564-020-0695-z PMid:32123347.
» http://dx.doi.org/10.1038/s41564-020-0695-z -
DE ALBUQUERQUE, N., BAIG, E., MA, X., ZHANG, J., HE, W., ROWE, A., HABAL, M., LIU, M., SHALEV, I., DOWNEY, G.P., GORCZYNSKI, R., BUTANY, J., LEIBOWITZ, J., WEISS, S.R., MCGILVRAY, I.D., PHILLIPS, M.J., FISH, E.N. and LEVY, G.A., 2006. Murine hepatitis virus strain 1 produces a clinically relevant model of severe acute respiratory syndrome in A/J mice. Journal of Virology, vol. 80, no. 21, pp. 10382-10394. http://dx.doi.org/10.1128/JVI.00747-06 PMid:17041219.
» http://dx.doi.org/10.1128/JVI.00747-06 -
FELSENSTEIN, J., 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution; International Journal of Organic Evolution, vol. 39, no. 4, pp. 783-791. http://dx.doi.org/10.1111/j.1558-5646.1985.tb00420.x PMid:28561359.
» http://dx.doi.org/10.1111/j.1558-5646.1985.tb00420.x -
GOMAA, M.H., BARTA, J.R., OJKIC, D. and YOO, D., 2008. Complete genomic sequence of turkey coronavirus. Virus Research, vol. 135, no. 2, pp. 237-246. http://dx.doi.org/10.1016/j.virusres.2008.03.020 PMid:18468711.
» http://dx.doi.org/10.1016/j.virusres.2008.03.020 - GRENINGER, A.L., PEPPER, G., SHEAN, R.C., CENT, A., PALILEO, I., KUYPERS, J.M., SCHIFFER, J.T. and JEROME, K.R., 2017. Myeloablation-associated deletion of ORF4 in a human coronavirus 229E infection. NPJ Genomic Medicine, vol. 2, no. 1, pp. 1-4.
-
HAN, Y., DU, J., SU, H., ZHANG, J., ZHU, G., ZHANG, S., WU, Z. and JIN, Q., 2019. Identification of diverse bat alphacoronaviruses and betacoronaviruses in China provides new insights into the evolution and origin of coronavirus-related diseases. Frontiers in Microbiology, vol. 10, pp. 1900. http://dx.doi.org/10.3389/fmicb.2019.01900 PMid:31474969.
» http://dx.doi.org/10.3389/fmicb.2019.01900 -
HEATH, L., VAN DER WALT, E., VARSANI, A. and MARTIN, D.P., 2006. Recombination patterns in aphthoviruses mirror those found in other picornaviruses. Journal of Virology, vol. 80, no. 23, pp. 11827-11832. http://dx.doi.org/10.1128/JVI.01100-06 PMid:16971423.
» http://dx.doi.org/10.1128/JVI.01100-06 -
HU, B., ZENG, L.P., YANG, X.L., GE, X.Y., ZHANG, W., LI, B., XIE, J.Z., SHEN, X.R., ZHANG, Y.Z., WANG, N., LUO, D.S., ZHENG, X.S., WANG, M.N., DASZAK, P., WANG, L.F., CUI, J. and SHI, Z.L., 2017. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathogens, vol. 13, no. 11, pp. e1006698. http://dx.doi.org/10.1371/journal.ppat.1006698 PMid:29190287.
» http://dx.doi.org/10.1371/journal.ppat.1006698 -
HU, D., ZHU, C., AI, L., HE, T., WANG, Y., YE, F., YANG, L., DING, C., ZHU, X., LV, R., ZHU, J., HASSAN, B., FENG, Y., TAN, W. and WANG, C., 2018. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerging Microbes & Infections, vol. 7, no. 1, pp. 1-10. http://dx.doi.org/10.1038/s41426-018-0155-5 PMid:30209269.
» http://dx.doi.org/10.1038/s41426-018-0155-5 -
HUANG, C., LIU, W.J., XU, W., JIN, T., ZHAO, Y., SONG, J., SHI, Y., JI, W., JIA, H., ZHOU, Y., WEN, H., ZHAO, H., LIU, H., LI, H., WANG, Q., WU, Y., WANG, L., LIU, D., LIU, G., YU, H., HOLMES, E.C., LU, L. and GAO, G.F., 2016. A bat-derived putative cross-family recombinant coronavirus with a reovirus gene. PLoS Pathogens, vol. 12, no. 9, pp. e1005883. http://dx.doi.org/10.1371/journal.ppat.1005883 PMid:27676249.
» http://dx.doi.org/10.1371/journal.ppat.1005883 -
HUSSAIN, S., PAN, J., CHEN, Y., YANG, Y., XU, J., PENG, Y., WU, Y., LI, Z., ZHU, Y., TIEN, P. and GUO, D., 2005. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. Journal of Virology, vol. 79, no. 9, pp. 5288-5295. http://dx.doi.org/10.1128/JVI.79.9.5288-5295.2005 PMid:15827143.
» http://dx.doi.org/10.1128/JVI.79.9.5288-5295.2005 - JI, W., WANG, W., ZHAO, X., ZAI, J. and LI, X., 2020. Homologous recombination within the spike glycoprotein of the newly identified coronavirus 2019-nCoV may boost cross-species transmission from snake to human. Journal of Medical Virology, vol. 22, no. 1, pp. 1-8.
-
JIN, Y.H., CAI, L., CHENG, Z.S., CHENG, H., DENG, T., FAN, Y.P., FANG, C., HUANG, D., HUANG, L.Q., HUANG, Q., HAN, Y., HU, B., HU, F., LI, B.H., LI, Y.R., LIANG, K., LIN, L.K., LUO, L.S., MA, J., MA, L.L., PENG, Z.Y., PAN, Y.B., PAN, Z.Y., REN, X.Q., SUN, H.M., WANG, Y., WANG, Y.Y., WENG, H., WEI, C.J., WU, D.F., XIA, J., XIONG, Y., XU, H.B., YAO, X.M., YUAN, Y.F., YE, T.S., ZHANG, X.C., ZHANG, Y.W., ZHANG, Y.G., ZHANG, H.M., ZHAO, Y., ZHAO, M.J., ZI, H., ZENG, X.T., WANG, Y.Y. and WANG, X.H., and for the Zhongnan Hospital of Wuhan University Novel Coronavirus Management and Research Team, Evidence-Based Medicine Chapter of China International Exchange and Promotive Association for Medical and Health Care (CPAM), 2020. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus [2019-nCoV] infected pneumonia. Military Medical Research, vol. 7, no. 1, pp. 4. http://dx.doi.org/10.1186/s40779-020-0233-6 PMid:32029004.
» http://dx.doi.org/10.1186/s40779-020-0233-6 - JUKES, T.H. and CANTOR, C.R., 1969. Evolution of protein molecules. In: H.N. MUNRO, ed. Mammalian protein metabolism New York: Academic Press, pp. 21-132. http://dx.doi.org/10.1016/B978-1-4832-3211-9.50009-7.
-
KELLEY, L.A., MEZULIS, S., YATES, C.M., WASS, M.N. and STERNBERG, M.J., 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols, vol. 10, no. 6, pp. 845-858. http://dx.doi.org/10.1038/nprot.2015.053 PMid:25950237.
» http://dx.doi.org/10.1038/nprot.2015.053 - LAU, S.K., WOO, P.C., LI, K.S., TSANG, A.K., FAN, R.Y., LUK, H.K., CAI, J.P., CHAN, K.H., ZHENG, B.J., WANG, M. and YUEN, K.Y., 2015. Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage. Journal of Virology, vol. 89, no. 6, pp. 3076-3092. PMid:25552712.
-
LI, F., 2016. Structure, function, and evolution of coronavirus spike proteins. Annual Review of Virology, vol. 3, no. 1, pp. 237-261. http://dx.doi.org/10.1146/annurev-virology-110615-042301 PMid:27578435.
» http://dx.doi.org/10.1146/annurev-virology-110615-042301 -
LIN, X.D., WANG, W., HAO, Z.Y., WANG, Z.X., GUO, W.P., GUAN, X.Q., WANG, M.R., WANG, H.W., ZHOU, R.H., LI, M.H., TANG, G.P., WU, J., HOLMES, E.C. and ZHANG, Y.Z., 2017. Extensive diversity of coronaviruses in bats from China. Virology, vol. 507, pp. 1-10. http://dx.doi.org/10.1016/j.virol.2017.03.019 PMid:28384506.
» http://dx.doi.org/10.1016/j.virol.2017.03.019 -
LU, H., STRATTON, C.W. and TANG, Y.W., 2020. Outbreak of pneumonia of unknown etiology in Wuhan China: the Mystery and the Miracle. Journal of Medical Virology, vol. 92, no. 4, pp. 401-402. http://dx.doi.org/10.1002/jmv.25678 PMid:31950516.
» http://dx.doi.org/10.1002/jmv.25678 -
MAGIORKINIS, G., MAGIORKINIS, E., PARASKEVIS, D., VANDAMME, A.M., VAN RANST, M., MOULTON, V. and HATZAKIS, A., 2004. Phylogenetic analysis of the full‐length SARS‐CoV sequences: evidence for phylogenetic discordance in three genomic regions. Journal of Medical Virology, vol. 74, no. 3, pp. 369-372. http://dx.doi.org/10.1002/jmv.20187 PMid:15368527.
» http://dx.doi.org/10.1002/jmv.20187 -
MARTIN, D.P., MURRELL, B., GOLDEN, M., KHOOSAL, A. and MUHIRE, B., 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evolution, vol. 1, no. 1, pp. vev003. http://dx.doi.org/10.1093/ve/vev003 PMid:27774277.
» http://dx.doi.org/10.1093/ve/vev003 -
MASTERS, P.S., 2006. The molecular biology of coronaviruses. Advances in Virus Research, vol. 66, pp. 193-292. http://dx.doi.org/10.1016/S0065-3527(06)66005-3 PMid:16877062.
» http://dx.doi.org/10.1016/S0065-3527(06)66005-3 -
MIHINDUKULASURIYA, K.A., WU, G., ST LEGER, J., NORDHAUSEN, R.W. and WANG, D., 2008. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. Journal of Virology, vol. 82, no. 10, pp. 5084-5088. http://dx.doi.org/10.1128/JVI.02722-07 PMid:18353961.
» http://dx.doi.org/10.1128/JVI.02722-07 -
NATIONAL CENTER FOR BIOTECHNOLOGY INFORMATION - NCBI [online], 2021 [viewed 11 August 2021]. Retrived from: http://www.ncbi.nlm.nih.gov
» http://www.ncbi.nlm.nih.gov -
OUR WORLD IN DATA, 2021 [viewed 11 August 2021]. Coronavirus (COVID-19) Vaccinations [online]. Retrived from: https://ourworldindata.org/covid-vaccinations
» https://ourworldindata.org/covid-vaccinations -
PALACIOS, G., DRUCE, J., DU, L., TRAN, T., BIRCH, C., BRIESE, T., CONLAN, S., QUAN, P.L., HUI, J., MARSHALL, J., SIMONS, J.F., EGHOLM, M., PADDOCK, C.D., SHIEH, W.J., GOLDSMITH, C.S., ZAKI, S.R., CATTON, M. and LIPKIN, W.I., 2008. A new arenavirus in a cluster of fatal transplant-associated diseases. The New England Journal of Medicine, vol. 358, no. 10, pp. 991-998. http://dx.doi.org/10.1056/NEJMoa073785 PMid:18256387.
» http://dx.doi.org/10.1056/NEJMoa073785 -
PARASKEVIS, D., KOSTAKI, E.G., MAGIORKINIS, G., PANAYIOTAKOPOULOS, G., SOURVINOS, G. and TSIODRAS, S., 2020. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infection, Genetics and Evolution, vol. 79, pp. 104212. http://dx.doi.org/10.1016/j.meegid.2020.104212 PMid:32004758.
» http://dx.doi.org/10.1016/j.meegid.2020.104212 -
PERLMAN, S. and NETLAND, J., 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nature Reviews. Microbiology, vol. 7, no. 6, pp. 439-450. http://dx.doi.org/10.1038/nrmicro2147 PMid:19430490.
» http://dx.doi.org/10.1038/nrmicro2147 -
RUSSELL, C.D., MILLAR, J.E. and BAILLIE, J.K., 2020. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet, vol. 395, no. 10223, pp. 473-475. http://dx.doi.org/10.1016/S0140-6736(20)30317-2 PMid:32043983.
» http://dx.doi.org/10.1016/S0140-6736(20)30317-2 - SAITOU, N. and NEI, M., 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, vol. 4, no. 4, pp. 406-425. PMid:3447015.
-
SINGHAL, T., 2020. A review of coronavirus disease-2019 (COVID-19). Indian Journal of Pediatrics, vol. 87, no. 4, pp. 281. http://dx.doi.org/10.1007/s12098-020-03263-6 PMid:32166607.
» http://dx.doi.org/10.1007/s12098-020-03263-6 -
TAKIAN, A., RAOOFI, A. and KAZEMPOUR-ARDEBILI, S., 2020. COVID-19 battle during the toughest sanctions against Iran. Lancet, vol. 395, no. 10229, pp. 1035-1036. http://dx.doi.org/10.1016/S0140-6736(20)30668-1 PMid:32199073.
» http://dx.doi.org/10.1016/S0140-6736(20)30668-1 -
TAMURA, K., STECHER, G., PETERSON, D., FILIPSKI, A. and KUMAR, S., 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution, vol. 30, no. 12, pp. 2725-2729. http://dx.doi.org/10.1093/molbev/mst197 PMid:24132122.
» http://dx.doi.org/10.1093/molbev/mst197 -
THIEL, V., HEROLD, J., SCHELLE, B. and SIDDELL, S.G., 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. The Journal of General Virology, vol. 82, no. Pt 6, pp. 1273-1281. http://dx.doi.org/10.1099/0022-1317-82-6-1273 PMid:11369870.
» http://dx.doi.org/10.1099/0022-1317-82-6-1273 -
VAN BOHEEMEN, S., DE GRAAF, M., LAUBER, C., BESTEBROER, T.M., RAJ, V.S., ZAKI, A.M., OSTERHAUS, A.D., HAAGMANS, B.L., GORBALENYA, A.E., SNIJDER, E.J. and FOUCHIER, R.A., 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio, vol. 3, no. 6, pp. 00473-12. http://dx.doi.org/10.1128/mBio.00473-12 PMid:23170002.
» http://dx.doi.org/10.1128/mBio.00473-12 -
VAN DER HOEK, L., PYRC, K., JEBBINK, M.F., VERMEULEN-OOST, W., BERKHOUT, R.J., WOLTHERS, K.C., WERTHEIM-VAN DILLEN, P.M., KAANDORP, J., SPAARGAREN, J. and BERKHOUT, B., 2004. Identification of a new human coronavirus. Nature Medicine, vol. 10, no. 4, pp. 368-373. http://dx.doi.org/10.1038/nm1024 PMid:15034574.
» http://dx.doi.org/10.1038/nm1024 -
VARSANI, A., SHEPHERD, D.N., MONJANE, A.L., OWOR, B.E., ERDMANN, J.B., RYBICKI, E.P., PETERSCHMITT, M., BRIDDON, R.W., MARKHAM, P.G., OLUWAFEMI, S., WINDRAM, O.P., LEFEUVRE, P., LETT, J.M. and MARTIN, D.P., 2008. Recombination, decreased host specificity and increased mobility may have driven the emergence of Maize streak virus as an agricultural pathogen. The Journal of General Virology, vol. 89, no. Pt 9, pp. 2063-2074. http://dx.doi.org/10.1099/vir.0.2008/003590-0 PMid:18753214.
» http://dx.doi.org/10.1099/vir.0.2008/003590-0 -
VIJGEN, L., KEYAERTS, E., MOËS, E., THOELEN, I., WOLLANTS, E., LEMEY, P., VANDAMME, A.M. and VAN RANST, M., 2005. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. Journal of Virology, vol. 79, no. 3, pp. 1595-1604. http://dx.doi.org/10.1128/JVI.79.3.1595-1604.2005 PMid:15650185.
» http://dx.doi.org/10.1128/JVI.79.3.1595-1604.2005 -
WANG, D., HU, B., HU, C., ZHU, F., LIU, X., ZHANG, J., WANG, B., XIANG, H., CHENG, Z., XIONG, Y. and ZHAO, Y., 2020. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China: JAMA. http://dx.doi.org/10.1001/jama.2020.1585
» http://dx.doi.org/10.1001/jama.2020.1585 - WANG, Y., SUN, J., ZHU, A., ZHAO, J. and ZHAO, J., 2018. Current understanding of Middle East respiratory syndrome coronavirus infection in human and animal models. Journal of Thoracic Disease, vol. 10, no. suppl. 19, pp. S2260.
-
WOO, P.C., LAU, S.K., CHU, C.M., CHAN, K.H., TSOI, H.W., HUANG, Y., WONG, B.H., POON, R.W., CAI, J.J., LUK, W.K., POON, L.L., WONG, S.S., GUAN, Y., PEIRIS, J.S. and YUEN, K.Y., 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. Journal of Virology, vol. 79, no. 2, pp. 884-895. http://dx.doi.org/10.1128/JVI.79.2.884-895.2005 PMid:15613317.
» http://dx.doi.org/10.1128/JVI.79.2.884-895.2005 -
WOO, P.C., LAU, S.K., LAM, C.S., LAI, K.K., HUANG, Y., LEE, P., LUK, G.S., DYRTING, K.C., CHAN, K.H. and YUEN, K.Y., 2009. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. Journal of Virology, vol. 83, no. 2, pp. 908-917. http://dx.doi.org/10.1128/JVI.01977-08 PMid:18971277.
» http://dx.doi.org/10.1128/JVI.01977-08 -
WOO, P.C., LAU, S.K., LAM, C.S., LAU, C.C., TSANG, A.K., LAU, J.H., BAI, R., TENG, J.L., TSANG, C.C., WANG, M., ZHENG, B.J., CHAN, K.H. and YUEN, K.Y., 2012. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. Journal of Virology, vol. 86, no. 7, pp. 3995-4008. http://dx.doi.org/10.1128/JVI.06540-11 PMid:22278237.
» http://dx.doi.org/10.1128/JVI.06540-11 -
WOO, P.C., WANG, M., LAU, S.K., XU, H., POON, R.W., GUO, R., WONG, B.H., GAO, K., TSOI, H.W., HUANG, Y., LI, K.S., LAM, C.S., CHAN, K.H., ZHENG, B.J. and YUEN, K.Y., 2007. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. Journal of Virology, vol. 81, no. 4, pp. 1574-1585. http://dx.doi.org/10.1128/JVI.02182-06 PMid:17121802.
» http://dx.doi.org/10.1128/JVI.02182-06 - WU, F., ZHAO, S., YU, B., CHEN, Y.M., WANG, W., HU, Y., SONG, Z.G., TAO, Z.W., TIAN, J.H., PEI, Y.Y. and YUAN, M.L., 2020. A novel coronavirus associated with a respiratory disease in Wuhan of Hubei province China: NCBI.
-
WU, Q., ZHANG, Y., LÜ, H., WANG, J., HE, X., LIU, Y., YE, C., LIN, W., HU, J., JI, J., XU, J., YE, J., HU, Y., CHEN, W., LI, S., WANG, J., WANG, J., BI, S. and YANG, H., 2003. The E protein is a multifunctional membrane protein of SARS-CoV. Genomics, Proteomics & Bioinformatics, vol. 1, no. 2, pp. 131-144. http://dx.doi.org/10.1016/S1672-0229(03)01017-9 PMid:15626343.
» http://dx.doi.org/10.1016/S1672-0229(03)01017-9 -
WU, Z., YANG, L., REN, X., HE, G., ZHANG, J., YANG, J., QIAN, Z., DONG, J., SUN, L., ZHU, Y., DU, J., YANG, F., ZHANG, S. and JIN, Q., 2016a. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. The ISME Journal, vol. 10, no. 3, pp. 609-620. http://dx.doi.org/10.1038/ismej.2015.138 PMid:26262818.
» http://dx.doi.org/10.1038/ismej.2015.138 -
WU, Z., YANG, L., REN, X., ZHANG, J., YANG, F., ZHANG, S. and JIN, Q., 2016b. ORF8-related genetic evidence for Chinese horseshoe bats as the source of human severe acute respiratory syndrome coronavirus. The Journal of Infectious Diseases, vol. 213, no. 4, pp. 579-583. http://dx.doi.org/10.1093/infdis/jiv476 PMid:26433221.
» http://dx.doi.org/10.1093/infdis/jiv476 -
XU, X.W., WU, X.X., JIANG, X.G., XU, K.J., YING, L.J., MA, C.L., LI, S.B., WANG, H.Y., ZHANG, S., GAO, H.N., SHENG, J.F., CAI, H.L., QIU, Y.Q. and LI, L.J., 2020. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-CoV-2) outside of Wuhan, China: retrospective case series. BMJ (Clinical Research Ed.), vol. 368, pp. 368. http://dx.doi.org/10.1136/bmj.m606 PMid:32075786.
» http://dx.doi.org/10.1136/bmj.m606 -
YANG, X.L., HU, B., WANG, B., WANG, M.N., ZHANG, Q., ZHANG, W., WU, L.J., GE, X.Y., ZHANG, Y.Z., DASZAK, P., WANG, L.F. and SHI, Z.L., 2016. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. Journal of Virology, vol. 90, no. 6, pp. 3253-3256. http://dx.doi.org/10.1128/JVI.02582-15 PMid:26719272.
» http://dx.doi.org/10.1128/JVI.02582-15 -
ZHANG, J., LIU, Y., HU, L., GAO, Q., ZHANG, Z., ZHANG, X., CHEN, J., GONG, X., SONG, L., LIU, Y., LI, J., LI, S., HUANG, J., NING, Y., GAO, H., QIN, C., DONG, X., WEI, J., DONG, G. and YIN, W., 2005. Preparation and characterization of SARS in-house reference antiserum. Vaccine, vol. 23, no. 48-49, pp. 5666-5669. http://dx.doi.org/10.1016/j.vaccine.2004.12.032 PMid:16269206.
» http://dx.doi.org/10.1016/j.vaccine.2004.12.032 -
ZHANG, L. and LIU, Y., 2020. Potential interventions for novel coronavirus in China: a systematic review. Journal of Medical Virology, vol. 92, no. 5, pp. 479-490. http://dx.doi.org/10.1002/jmv.25707 PMid:32052466.
» http://dx.doi.org/10.1002/jmv.25707 - ZHAO, J.P., HU, Y., DU, R.H., CHEN, Z.S., JIN, Y., ZHOU, M., ZHANG, J., QU, J.M. and CAO, B., 2020. Expert consensus on the use of corticosteroid in patients with 2019-nCoV pneumonia. Zhonghua Jie He He Hu Xi Za Zhi, vol. 43, no. 3, pp. 183-184. PMid:32164084.
- ZHENG, H., WEI, C., DING, R., WANG, W., LI, W., WANG, J., TAO, W., YU, X., GUO, H., CHEN, J., WEI, W., LI, J., ZHANG, Y., WANG, X., SUN, Y., JIAO, J., WANG, Y. and ZHOU, C., 2003. SARS coronavirus NS-1 complete genome sequence Unpublished.
-
ZHOU, P., YANG, X.L., WANG, X.G., HU, B., ZHANG, L., ZHANG, W., SI, H.R., ZHU, Y., LI, B., HUANG C.L., CHEN, H.D., CHEN, J., LUO, Y., GUO, H., JIANG, R.D., LIU, M.Q., CHEN, Y., SHEN, X., WANG, X., ZHENG, X.S., ZHAO, K., CHEN, Q.J., DENG, F., LIU, L.L., YAN, B., ZHAN, F.X., WANG, Y.Y., XIAO, G.F. and SHI, Z.L., 2020. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. BioRxiv http://dx.doi.org/10(2020.01):22-914952
» http://dx.doi.org/10(2020.01):22-914952 - ZHOU, X., HOU, J., HE, M., DING, Y., WANG, Z., LI, J., LIU, Z., MA, S., ZHANG, K., PAN, W., ZHANG, J., HE, J., WANG, Y., XU, Y., and WANG, X., 2003. SARS coronavirus GZ02 isolate genome sequence EMBL/GenBank/DDBJ databases (Submitted (SEP-2003)).
-
ZHU, N., ZHANG, D., WANG, W., LI, X., YANG, B., SONG, J., ZHAO, X., HUANG, B., SHI, W., LU, R., NIU, P., ZHAN, F., MA, X., WANG, D., XU, W., WU, G., GAO, G.F. and TAN, W., and CHINA NOVEL CORONAVIRUS INVESTIGATING AND RESEARCH TEAM, 2020. A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, vol. 382, no. 8, pp. 727-733. http://dx.doi.org/10.1056/NEJMoa2001017 PMid:31978945.
» http://dx.doi.org/10.1056/NEJMoa2001017 -
ZIEBUHR, J., GORBALENYA, A.E. and SNIJDER, E.J., 2000. Virus‐encoded proteinases and proteolytic processing in the Nidovirales. The Journal of General Virology, vol. 81, no. Pt 4, pp. 853-879. http://dx.doi.org/10.1099/0022-1317-81-4-853 PMid:10725411.
» http://dx.doi.org/10.1099/0022-1317-81-4-853
Publication Dates
-
Publication in this collection
03 Sept 2021 -
Date of issue
2023
History
-
Received
05 Jan 2021 -
Accepted
09 Apr 2021

 Current understanding of an Emerging Coronavirus using in silico approach: Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2)
Current understanding of an Emerging Coronavirus using in silico approach: Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2)