Abstract
Sisyrinchium micranthum Cav. is a member of the family Iridaceae, which is distributed over the American continent. In Brazil, this species is found, not only in disturbed areas and coastal regions, but is also very common in urban centers, such as public parks, during the spring. Chromosome counts for North American specimens are 2n = 32 and 2n = 48, whereas in southern Brazil, there is a polyploidy series with three chromosome numbers, 2n = 16, 2n = 32, and 2n = 48. Population analyses using DNA molecular markers are inexistent for this species, in spite of its wide distribution and morphological variation. To study the genetic population structure of S. micranthum, five natural populations were accessed in a conservation park within the Atlantic Rain Forest Biome in southern Brazil. Here, the chromosome numbers 2n = 16 and 2n = 48 had already been described. Molecular analysis showed that the populations are highly structured with low gene flow among them. The population with 2n = 48 was genetically less variable than and distinct from the other populations. Population genetics in relation to cytogenetic data provided new insights regarding the genetic diversification and mating system of S. micranthum.
Iridaceae; ISSR-PCR; mating system; population genetics; Sisyrinchium micranthum
Population genetic structure of Sisyrinchium micranthum Cav. (Iridaceae) in Itapuã State Park, Southern Brazil
Luana Olinda TacuatiáI; Lilian EggersII; Eliane Kaltchuk-SantosI; Tatiana T. Souza-ChiesI, II
IInstituto de Biociências, Departamento de Genética, Programa de Pós-Graduação em Genética e Biologia Molecular, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brazil
IIInstituto de Biociências, Departamento de Botânica, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brazil
Send correspondence to Send correspondence to: Luana Olinda Tacuatiá Instituto de Biociências Departamento de Genética Programa de Pós-Graduação em Genética e Biologia Molecular Universidade Federal do Rio Grande do Sul Avenida Bento Gonçalves 9500, Prédio 43312, Caixa Postal 15053 91501-970 Porto Alegre, RS, Brazil E-mail: tacuatia@yahoo.com.br
ABSTRACT
Sisyrinchium micranthum Cav. is a member of the family Iridaceae, which is distributed over the American continent. In Brazil, this species is found, not only in disturbed areas and coastal regions, but is also very common in urban centers, such as public parks, during the spring. Chromosome counts for North American specimens are 2n = 32 and 2n = 48, whereas in southern Brazil, there is a polyploidy series with three chromosome numbers, 2n = 16, 2n = 32, and 2n = 48. Population analyses using DNA molecular markers are inexistent for this species, in spite of its wide distribution and morphological variation. To study the genetic population structure of S. micranthum, five natural populations were accessed in a conservation park within the Atlantic Rain Forest Biome in southern Brazil. Here, the chromosome numbers 2n = 16 and 2n = 48 had already been described. Molecular analysis showed that the populations are highly structured with low gene flow among them. The population with 2n = 48 was genetically less variable than and distinct from the other populations. Population genetics in relation to cytogenetic data provided new insights regarding the genetic diversification and mating system of S. micranthum.
Key words: Iridaceae, ISSR-PCR, mating system, population genetics, Sisyrinchium micranthum.
Introduction
Sisyrinchium micranthum Cav. (Iridaceae) is an herb species with violet, yellow, or pink flowers, violet being the most common color. This species produces floral oil in trichomatic structures called elaiophores, as a reward to pollinators (Cocucci and Vogel, 2001; Truylio et al., 2002). S. micranthum is distributed in Americas (Johnston, 1938; Goldblatt, 2003), from south Argentina to Mexico. In Brazil, it is usually encountered in disturbed areas, and during the spring, it is commonly to be found flowering in urban centers, such as public parks. In south Brazil, this herb shows remarkable morphological variation, and different morphological categories (CI, CII, and CIII) have been adopted to classify plants based on morphological features, such as the number of internodes of the flowering stem, as well as the lengths of the flowering stem, the inferior internode, the peduncle, the outer and inner spathes and the staminal column (Tacuatiá LO, Flores AM, Souza-Chies TT, Eggers L, Siljak-Yakovlev S, Kaltchuk-Santos E, submitted).
Iridaceae is represented by around 65 to 75 genera, with over 2030 species all told (Goldblatt et al., 2008). Certain genera have been studied extensively, due to their economic importance as ornamental plants, food items and spices. Little is known, however, regarding most of those devoid of economic value, such as Sisyrinchium L. species. This genus is represented by approximately 140 species in America (Goldblatt et al., 2008). Data on the biology, cytogenetics, and leaf anatomy of Sisyrinchium species are available, especially for North American species (Ingram, 1968; Henderson, 1976; Cholewa and Henderson, 1984; Goldblatt et al., 1984; Kenton et al., 1986; Rudall et al., 1986; Goldblatt and Takei, 1997), although little is known about most of those from South America, especially from Brazil.
Iridaceae species, including Iris L., Moraea Mill., Crocus L., Gladiolus L. and the genus Sisyrinchium, have extensive polyploid series (Ingram, 1968; Henderson, 1976; Cholewa and Henderson, 1984; Goldblatt et al., 1984; Kenton and Heywood, 1984; Kenton et al., 1986; Rudall et al., 1986; Goldblatt and Takei, 1997). Polyploidy, reported in more than 70% of Sisyrinchium species (Goldblatt and Takei, 1997), also appears to be related to the complex diversification of S. micranthum (Tacuatiá LO, Flores AM, Souza-Chies TT, Eggers L, Siljak-Yakovlev S, Kaltchuk-Santos E, submitted). Goldblatt (1982) described the chromosome number for introduced specimens collected in Texas as 2n = 32, whereas native plants from Colombia (Kenton and Heywood, 1984) and Nicaragua (Goldblatt and Takei, 1997) presented 2n = 48. In south Brazil, three cytotypes (2x, 4x and 6x) have recently been recorded for S. micranthum, diploidy (2n = 16) being the most common (Tacuatiá LO, Flores AM, Souza-Chies TT, Eggers L, Siljak-Yakovlev S, Kaltchuk-Santos E, submitted). The allopolyploid origin of many cytotypes in the genus Sisyrinchium is proposed by Kenton et al. (1986), although consistent data are not available.
The population genetics of Iridaceae species have been intensively investigated (Burke and Arnold, 1999; Burke et al., 2000; Hannan and Orick, 2000; Wilson et al., 2000; Karst and Wilson, 2002; Wróblewska et al., 2003; Caiola et al., 2004; Meerow et al., 2005, 2007; Marco et al., 2009). Even so, and despite its wide distribution and interesting morphological and genetic features, S. micranthum has been neglected. The Itapuã State Park (Parque Estadual de Itapuã-PEI), where several populations of different morphological categories have been observed, is a state conservation area, dedicated to preserving the remaining original vegetation and impressive ecosystem diversity of the Atlantic Rain Forest Biome (Fonseca et al., 2004).
The present study aims to investigate genetic variability within and among these populations by means of inter-simple sequence repeat (ISSR) markers.
Material and Methods
Population sampling
In 2005, five populations of S. micranthum were sampled in the PEI located in the municipality of Viamão, approximately 57 km from Porto Alegre, Rio Grande do Sul (RS), Brazil (Figure 1). The study sites consisted of a bank site, denominated Guaíba Lagoon, and sites comprising hills with granitic outcrops, viz., Praia de Fora, Pedra da Visão, Pedra da Grota and Praia da Pedreira. Four populations with light violet flowers and one with two flower colors, light violet and light yellow, were collected. Collection sites, coordinates, numbers of collected individuals, and flower colors appear in Table 1. Voucher specimens were deposited in the ICN Herbarium, Instituto de Biociências, Universidade Federal do Rio Grande do Sul.
DNA isolation and ISSR-PCR amplification
DNA sample extraction was based on the method of Doyle and Doyle (1987) with certain modifications. A set of twelve ISSR primers was tested, and six that generated good patterns with a representative sample group were further used for DNA amplification of all the populations. PCR was carried out in 25-µL reactions using (depending on the primer): 4% DMSO, 1x buffer, 4.6-5.0 mM MgCl2, 0.48-0.8 mM dNTP mixture (Invitrogen, São Paulo, Brazil), 0.4-0.6 mM of each primer, 1 U Taq DNA polymerase (CenBiot, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brazil), and 10 ng of genomic DNA. The thermal cycling program for amplification consisted of initial denaturation at 92 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 42-45 °C (Table 2) for 1 min, extension at 72 °C for 2 min 30 s, and a final extension step at 72 °C for 5 min. PCR products were analyzed on 1.5% agarose gels and stained with GelRed (Amicon Corp., Lexington, MA).
Statistical analyses
Bands were scored as binary characters based on their presence (1) or absence (0) and an unbiased genetic distance matrix (Nei, 1978) was generated by TFPGA version 1.3 (Tools for Population Genetic Analyses; Miller, 1997) to construct an unweighted pair-group method arithmetic average (UPGMA) topology, which computed 1000 permutations and estimated the confidence limits of the dendrogram. Marker frequencies were estimated based on the Lynch and Milligan (1994) Taylor expansion estimate. One pairwise difference matrix generated by ARLEQUIN version 3.11 (Excoffier et al., 2005) with 1000 permutations, was used with MEGA version 4.0 (Tamura et al., 2007) to produce an UPGMA dendrogram.
To test the correlation between the pairwise ΦST matrix generated by ARLEQUIN and the unbiased genetic distance matrix generated by TFPGA, and between genetic and geographic distances (in km) among populations, a Mantel Test was performed using TFPGA with 10,000 permutations.
A hierarchical analysis of molecular variance (AMOVA; Excoffier et al., 1992) was obtained with ARLEQUIN version 3.11 to determine the variance components and their significance levels.
Considering Bayesian analyses, a Bayesian approach proposed by Holsinger et al. (2002) was also applied with HICKORY version 1.1, to obtain a more direct estimate of FST from dominant markers, unaffected by Hardy-Weinberg and f assumptions. The a posteriori distribution of the θB estimator (the estimate of FST) was numerically approximated through a Markov Chain Monte Carlo (MCMC) simulation, and tends to converge to a beta distribution (see Telles et al., 2006, for a better application). The four models available in the software were tested, i.e., the full model which allows estimating both θB and f, models with f or θB equal to zero, and, a final model leaving f free to vary so that the sampler does not attempt to estimate f, but chooses f values from its prior distribution, while estimating other parameters during the MCMC run. Model choice was based on the Deviance Information Criterion (DIC; Spiegelhalter et al., 2002). Estimates of genetic diversity (hs; defined as average panmictic heterozygosity) within each population were also calculated.
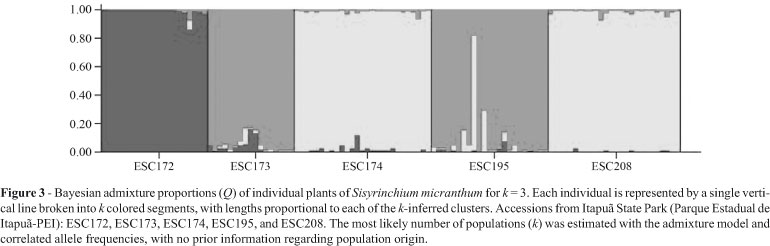
STRUCTURE version 2.3.1 (Falush et al., 2007) was employed to obtain additional insights regarding gene flow and population subdivision. The most likely number of populations (k) was estimated under the admixture model and correlated allele frequencies, with no prior information on population origin. The program was run for 10,000 iterations, after a burn-in length of 10,000 iterations, to test population subdivision from k = 1 to k = 10, and thereby check for any possible subdivision. Twenty runs were carried out for each k, to quantify variation in likelihood, as a means of checking whether different runs could produce different likelihood values. Individual and average admixture proportions (Q) for each population in each genetic cluster found by the program, were recorded for the model. As an aid in identifying the number of clusters of individuals (k), the results generated by STRUCTURE were subsequently analyzed by way of the STRUCTURE HARVESTER version 0.6.7 (Earl, 2011), according to the method of Evanno et al. (2005). This method uses an ad hoc statistic k, based on the rate of change in the log probability of data between successive k values (see Evanno et al., 2005, for a better explanation).
Results
The six primers produced 80 computable bands, of which 98.75% were polymorphic. The ISSR fragments generated an average of 13.2 bands per primer. The size of the amplified products ranged from 325 to 1800 bp (Table 2), the percentages of polymorphic loci from 43.8% (ESC172) to 78.8% (ESC195; Table 3), and genetic diversity indices within each population from 0.19 to 0.25 (results from Bayesian analysis in HICKORY, full model; Table 3).
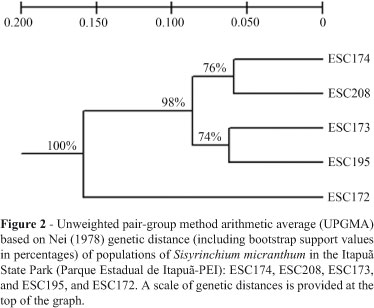
Both, the UPGMA produced by TFPGA (Figure 2) based on the Nei's unbiased genetic distance matrix, and the UPGMA dendrogram (not shown) generated by MEGA based on the ΦST pairwise difference matrix, presented two main clusters, one comprising only the ESC172 population and the second two subclusters (bootstrap value about 98%), the first subcluster consisting of the ESC173 and ESC195 populations, and the second of the ESC174 and ESC208 populations (74% and 76% of bootstrap values, respectively).
AMOVA generated Φ statistics, one analogous to Wright's F statistics. The analysis revealed that approximately 33% (ΦST = 0.3372, p < 0.001) of genetic diversity could be attributed to divergence among populations, and 66% between individuals within a population (ΦIS = 0.6628, p < 0.001).
The Mantel test showed no significant correlation between geographic and genetic distances (r = 0.2663, p = 0.2204).
Data obtained through Bayesian genetic-structure analyses of S. micranthum furnished additional insights into population differentiation and gene flow. According to population analysis performed with HICKORY, the best model, i.e., that which presented the lowest DIC (DIC = 1424.0), was the full model, with θB = 0.49 and f = 0.65. The second best was f = 0 (DIC = 1468.4), with θB = 0.43. Although in both cases, θB values were higher than the ΦST presented by AMOVA, all the FST analogues detected outstanding population differentiation. However, the values of inbreeding coefficients derived from analyses were very similar, ΦIS = 0.66 (ARLEQUIN, AMOVA) and f = 0.65 (HICKORY, full model). The f-free model presented f = 0.50 and θB = 0.49, the same result forθB presented by the full model, and DIC = 1582.0. The worst model of all wasθB = 0, resulting inf = 0.91, and DIC = 3853.5.
The k = 3 model was the most adequate for elucidating clustering (Figure 3). The decision was made based on statistic k, in so far as the uppermost peak of its modal value corresponded to the number of clusters detected by the software (Figure 4). The clustering into three groups corresponded exactly to the UPGMA produced by TFPGA (Figure 2); populations ESC174 and ESC208, and, ESC173 and ESC195 grouped together, and accession ESC172 represented apart in a third cluster (Figure 3).
Discussion
In spite of belonging to such a diversified family as Iridaceae, Sisyrinchium micranthum has never undergone population analysis using DNA molecular markers, whereby the importance of population genetics, not only in the specific case of diversity analysis, but also in bringing together other primary studies of this species. Based on Nei (1978) genetic distance, it was possible to cluster the different accessions into well-supported branches (Figure 2). The populations ESC173, ESC174, ESC195, and ESC208 were grouped into one of the two main clusters, and ESC172 into the other. Through previous cytogenetic analysis, the haploid chromosome numbers of three proved to be n = 8 (ESC173 and ESC208) and n = 24 (ESC172), corresponding to the somatic numbers 2n = 16 (diploid) and 2n = 48 (hexaploid), respectively (Tacuatiá LO, Flores AM, Souza-Chies TT, Eggers L, Siljak-Yakovlev S, Kaltchuk-Santos E, submitted). Since ESC195 and ESC174 clustered with ESC173 and ESC208, respectively, it is possible that both can be considered diploids.
The data obtained showed that the highly structured sites (ΦST = 0.3372, θB = 0.49) corresponded to different populations. Variance within populations, besides elucidating about 65%-66% (ΦIS = 0.66; and f = 0.65) of the total variation, corresponded well with the genetic structure of outcrossing plants. In general, there is more overall genetic variation and less differentiation among populations of outcrossing plants than in selfing plants (Hamrick and Godt, 1996). Truylio et al. (2002), when studying flower biology of S. micranthum in São Francisco de Paula, RS, in south Brazil, found that this species is protogynous, i.e., female flower receptivity begins 6 to 7 h before male maturation. Moreover, in the same study, controlled pollination experiments indicated self-incompatibility. The pollinators were oil-bees from the family Apidae, of the tribe Tapinotaspidini (Cocucci and Vogel, 2001; Truylio et al., 2002), which usually nest close to foraging areas. However, syrphids and small pollen-collecting bees have already been seen visiting the flowers of S. micranthum (Truylio et al., 2002; Freitas and Sazima, 2006). Although it is not clear how the plants investigated by Truylio et al. (2002) should be classified, as regards the morphological categories of S. micranthum adopted here, the low interpopulation gene flow verified in the present study might be related to pollinator behavior.
It has already been shown (Holtsford and Ellstrand, 1989; Martín et al., 1997) that among herb species there is a correlation between high levels of intrapopulational variability and the breeding system. When using isozymes, Holtsford and Ellstrand (1989) found that, in the annual herb Clarkia tembloriensis Vasek (Onagraceae), the breeding system exerts a strong influence upon the distribution of genetic variation, both within and among populations. In this case, outcrossing populations had more total genetic variation and lower levels of differentiation among populations than the group of selfing plants. Erodium paularense Fern. Gonz. & Izco (Geraniaceae) is an outbreeding species, endemic to Spain. Population genetics studies showed that in this species, about 80% of all genetic diversity can be attributed to intrapopulational variation, thus consistent with the population structure of allogamous plants as a whole (Martín et al., 1997).
It is important to note that in the ESC172 population, the percentage of polymorphic loci was lower, it was clustered separately from the other populations (Figure 2), in most individuals there was no admixture of alleles with those of the other populations (Figure 3), and the genetic diversity index was the lowest (Table 3). Thus, as there was no correlation between geographic and genetic distances; differentiation among collection sites remained enigmatic.
Since hexaploid plants (e.g., ESC172) belong to the Cl morphological type (reduced plant-size and anther-height, and larger pollen grains), according to the classification adopted for S. micranthum in south Brazil (Tacuatiá LO, Flores AM, Souza-Chies TT, Eggers L, Siljak-Yakovlev S, Kaltchuk-Santos E, submitted), the isolated clustering of the ESC172 population in the dendrogram might be related to the different chromosome numbers, thereby possibly associating polyploidy to this populational divergence.
These aspects raise questions as to whether the breeding system could be somehow related to genetic differentiation between ESC172 and the other populations analyzed, and polyploidy. Henderson (1976), when studying Sisyrinchium species from the Northern Hemisphere, reported a correlation between breeding system and ploidy level. Hand-selfing procedures showed that tetraploids were self-incompatible, whereas most of the higher polyploids were self-fertile. Furthermore, anthesis observations indicated that protandry (maturation of the anthers before the stigma) often occurred in blue-eyed grasses, thereupon inducing outcrossing, even in self-compatible plants. However, a higher ploidy level was associated with a shorter protandrous state, due to a decrease in the time interval between anther and stigma maturation according to an increase in ploidy level. Thus, while tetraploids were protandrous, most octoploids and dodecaploids presented a short or even no maturation time interval, thereby inducing higher levels of self-compatibility and self-pollination in plants with higher polyploidy.
Another interesting example is S. bermudiana, a species that occurs over a wide area of North America. This plant is self-fertile and presents a large range of chromosome numbers (2n = 32, 64, 96; Kenton et al., 1986; Ingram, 1968). Its flowers are protandrous, since the anthers dehisce before the flower opens, whereas the stigma matures after opening. Furthermore, the length of the filament column is variable. Consequently, self-pollination is highly probable when the anther is at the same level as the style. Even so, outcrossing may occur when the filament column is shorter than the style (Ingram, 1968). Although, in the case of S. bermudiana, the relationship between ploidy level and self-pollination has not been investigated, from a previous study of Sisyrinchium (Henderson, 1976), it appears that the variation in length of the filament column in S. bermudiana could be related to the variation in chromosome number.
Based on the low values of genetic diversity presented by the polyploid accession in the PEI (ESC172), and considering that polyploidy, as mentioned above, may result in changes in the breeding system, this low genetic variation could arise from selfing. Thus, the populations, ESC173, ESC174, ESC195, and ESC208 are presumably diploids, self-incompatible and mainly outcrossing, according to the reported literature and the high estimated genetic diversity. On the other hand, since ESC172 is a polyploid population and genetically less variable, this population is possibly composed of self-compatible and self-fertile individuals.
However, the protogynous condition remains to be considered. As in the other Sisyrinchium species reported, the polyploidy process at ESC172 population could have been instrumental in breaking down this characteristic and/or self-incompatibility. Thus, as outcrossing would no longer be favored, self-pollination could occur.
As five specimens of ESC172 (with light-violet-colored flowers) presented the same band profile, four were excluded from statistical analyses. This pattern suggests that they are the result of self-fertilization or may even be clones. Thus, asexual reproduction may be contributing to the maintenance of this population. This is the first indication that reproduction in this species may not be strictly sexual. Thus, if reproduction is also vegetative, how would this fit into the mating system? On studying the relationship between mating system and life history (annual/perennial) in Solanum L. (Solanaceae), Vallejo-Marín and O'Brien (2007) found self-incompatibility and clonality to be strongly correlated. In their study, all of the self-incompatible plants were clonal and all strict annuals were self-compatible. Even so, in Decodon verticillatus (L.) Elliott (Lythraceae), clonality potentially furthers the increase of selfed offspring by way of geitonogamy (selfing through pollen transfer between flowers on the same plant; Eckert, 2000). Thus, the low genetic diversity in ESC172 individuals may possibly arise from self-fertilization alone, or as a consequence of both clonality and selfing. The connection of life-history with these aspects is unclear, as S. micranthum is usually cited as annual (Johnston, 1938; Innes, 1985; Goldblatt, 2003). Nonetheless, Parent (1987) reported specimens in northwest Spain that lived for more than one year, or at least survived the winter.
Even though whether something similar is occurring or has already occurred, and for how long, in S. micranthum in the PEI is unknown, this comprises an interesting mechanism for population maintenance in cases of a pre- or post-zygotic barrier between hexaploids and diploids at the beginning of colonization. Thus, the lower level of differentiation among individuals in the ESC172 population may be a consequence of initial population formation through the combination of a few polyploid specimens and the mating system.
Additional issues have emerged from the data regarding diversification and reproductive and pollination biology in this plant species. In order to adequately address the remaining questions, the need arises for alternative approaches to elucidate the mechanisms involved in its diversity, as well as a better understanding of its biology as a whole.
Acknowledgments
The authors thank M Pinheiro, Prof. F Bered, C Palma-Silva, and Prof. MH Bodanese-Zanettini for assistance and suggestions, G Agostini, F Spier, and ÉG de Marco for laboratory assistance, and CNPq, FAPERGS, CAPES, Propesq/UFRGS for financial support.
Internet Resources
Received: June 14, 2011
Accepted: August 30, 2011.
Associate Editor: Everaldo Gonçalves de Barros
License information: This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- Burke JM and Arnold ML (1999) Isolation and characterization of microsatellites in Iris Mol Ecol 8:1075-1092.
- Burke JM, Bulger MR, Wesselingh RA and Arnold ML (2000) Frequency and spatial patterning of clonal reproduction in Louisiana Iris hybrid populations. Evolution 54:137-144.
- Caiola MG, Caputo P and Zanier R (2004) RAPD analysis in Crocus sativus L. accessions and related Crocus species. Biol Plant 48:375-380.
- Cholewa AF and Henderson DM (1984) Biosystematics of Sisyrinchium section Bermudiana (Iridaceae) of the Rocky Mountains. Brittonia 36:342-363.
- Cocucci AR and Vogel S (2001) Oil-producing flowers of Sisyrinchium species (Iridaceae) and their pollinators in southern South America. Flora 196:26-46.
- Doyle JJ and Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11-15.
- Eckert CG (2000) Contributions of autogamy and geitonogamy to self-fertilization in a mass-flowering, clonal plant. Ecology 81:532-542.
- Evanno G, Regnaut S and Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol 14:2611-2620.
- Excoffier L, Smouse PE and Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 131:479-491.
- Excoffier L, Laval G and Schneider S (2005) Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online 1:47-50.
- Falush D, Stephens M and Pritchard JK (2007) Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol Ecol 7:574-578.
- Freitas L and Sazima M (2006) Pollination biology in tropical high-altitude grassland in Brazil: Interactions at the community level. Ann Mo Bot Gard 93:465-516.
- Goldblatt P (1982) Chromosome cytology in relation to suprageneric systematics of Neotropical Iridaceae. Syst Bot 7:186-198.
- Goldblatt P (2003) Iridaceae. In: Hammel BE, Grayum MH, Herrera C and Zamora N (eds) Manual de plantas de Costa Rica II, Syst Bot Monogr v. 92. Missouri Botanical Garden Press, St. Louis, pp 603-612.
- Goldblatt P and Takei M (1997) Chromosome cytology of Iridaceae - Patterns of variation, determination of ancestral base numbers, and modes of karyotype change. Ann Mo Bot Gard 84:285-304.
- Goldblatt P, Walbot V and Zimmer EA (1984) Estimation of genome size (C-Value) in Iridaceae by cytophotometry. Ann Mo Bot Gard 71:176-180.
- Goldblatt P, Rodriguez A, Powell MP, Davies TJ, Manning JC, Van Der Bank M and Savolainen V (2008) Iridaceae "out of Australasia"? Phylogeny, biogeography, and divergence time based on plastid DNA sequences. Syst Bot 33:495-508.
- Hamrick JL and Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc Lond B Biol Sci 351:1291-1298.
- Hannan GL and Orick MW (2000) Isozyme diversity in Iris cristata and threatened glacial endemic I. lacustris (Iridaceae). Am J Bot 87:293-301.
- Henderson DM (1976) A biosystematic study of Pacific Northwestern blue-eyed grasses (Sisyrinchium, Iridaceae). Brittonia 28:149-176.
- Holsinger KE, Lewis PO and Dey DK (2002) A Bayesian method for analysis of genetic population structure with dominant marker data. Mol Ecol 11:1157-1164.
- Holtsford TP and Ellstrand NC (1989) Variation in outcrossing rate and population genetic structure of Clarkia tembloriensis (Onagraceae). Theor Appl Genet 78:480-488.
- Ingram R (1968) Breeding barriers in some species of Sisyrinchium New Phytol 67:197-204.
- Innes C (1985) The world of Iridaceae - a comprehensive record. Holly Gate International Ltd., Ashington, 405 pp.
- Johnston IM (1938) The species of Sisyrinchium in Uruguay, Paraguay and Brazil. J Arnold Arbor 19:376-401.
- Kenton A and Heywood CA (1984) Cytological studies in South American Iridaceae. Plant Syst Evol 146:87-104.
- Kenton AY, Rudall PJ and Johnson AR (1986) Genome size variation in Sisyrinchium L. (Iridaceae) and its relationship to phenotype and habitat. Bot Gaz 147:342-354.
- Lynch M and Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91-99.
- Marco EG de, Tacuatiá LO, Eggers L, Kaltchuk-Santos E and Souza-Chies TT (2009) Genetic variability within Cypella fucata Ravenna in southern Brazil. In: Mahoney CL and Springer DA (eds) Genetic Diversity. Nova Publishers, New York, pp 179-194.
- Martín JP and Sánchez-Yélamo MD (1997) Genetic diversity within and among populations of a threatened species Erodium paularense Fern.Gonz. & Izco. Mol Ecol 6:813-820.
- Meerow AW, Gideon M, Kuhn DN and Schnell RJ (2005) Isolation and characterization of 10 microsatellite loci from Iris hexagona (Iridaceae). Mol Ecol Notes 5:410-412.
- Meerow AW, Gideon M, Kuhn DN, Motamayor JC and Nakamura K (2007) genetic structure and gene flow among south Florida populations of Iris hexagona Walt. (Iridaceae) assessed with 19 microsatellite DNA loci. Int J Plant Sci 168:1291-1309.
- Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583-590.
- Parent GH (1987) Données chorologiques et écologiques nouvelles sur le genre Sisyrinchium L. (Iridaceae) en Europe, avec quelques considerations nomenclaturales. Lejeunia: Rev Bot 121:1-16.
- Rudall P, Kenton AY and Lawrence TJ (1986) An anatomical and chromosomal investigation of Sisyrinchium and allied genera. Bot Gaz 147:466-477.
- Spiegelhalter DJ, Best NG, Carlin BP and van der Linde A (2002) Bayesian measures of model complexity and fit. J R Stat Soc Series B Stat Methodol 64:483-689.
- Tamura C, Dudley J, Nei M and Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software ver. 4.0. Mol Biol Evol 24:1596-1599.
- Telles MPC, Bastos RP, Soares TN, Resende LV and Diniz-Filho JAF (2006) RAPD variation and population genetic structure of Physalaemus cuvieri (Anura, Leptodactylidae) in Central Brazil. Genetica 128:323-332.
- Truylio B, Harter-Marques B and Engels W (2002) Biologia floral e polinização de Sisyrinchium micranthum (Iridaceae) na região do planalto das araucárias do Rio Grande do Sul, Brasil. Biociências 10:11-24.
- Vallejo-Marín M and O'Brien HE (2007) Correlated evolution of self-incompatibility and clonal reproduction in Solanum (Solanaceae). New Phytol 173:415-421.
- Wilson BL, Doede DL and Hipkins VD (2000) Isozyme variation in Sisyrinchium sarmentosum (Iridaceae). Northwest Sci 74:342-354.
- Wróblewska A, Brzosko E, Czarnecka BE and Nowosielski J (2003) High levels of genetic diversity in populations of Iris aphylla L. (Iridaceae), an endangered species in Poland. Bot J Linn Soc 142:65-72.
- Earl DA (2011) Structure harvester, ver. 0.6.1. http://taylor0.biology.ucla.edu/structureHarvester (August 5, 2011).
- Fonseca GAB, Rylands A, Paglia A and Mittermeier RA (2004) Atlantic Forest. http://multimedia.conservation.org/cabs/online_pubs/hotspots2/Atlanticforest.html (February 12, 2008).
- Karst L and Wilson CA (2002) Genetic variation in the rare endemic Sisyrinchium sarmentosum (Iridaceae) based on RAPDs. Botanical Society of America. Proceedings p 646. http://2002.botanyconference.org/section12/abstracts/214.shtml (February 30, 2008).
- Miller MP (1997) Tools for Population Genetic Analyses (TFPGA), ver. 1.3. A windows program for the analysis of allozyme and molecular population genetic data. Computer software distributed by author. http://www.marksgeneticsoftware.net/_vti_bin/shtml.exe/tfpga.htm (January 10, 2008).
Publication Dates
-
Publication in this collection
02 Feb 2012 -
Date of issue
2012
History
-
Received
14 June 2011 -
Accepted
30 Aug 2011

 Population genetic structure of Sisyrinchium micranthum Cav. (Iridaceae) in Itapuã State Park, Southern Brazil
Population genetic structure of Sisyrinchium micranthum Cav. (Iridaceae) in Itapuã State Park, Southern Brazil