Abstract
Hepatitis A virus (HAV) infection is a public health problem worldwide and the virus has been classified into six genotypes. In Brazil, the only genotype that has been found is genotype I, predominately from subgenotype IA. Here, the HAV genotypes were analyzed of 18 isolates circulating between 1996-2001 in Goiânia, state of Goiás, Brazil. Viral RNA was extracted from 18 serum samples and amplified (RT-PCR/nested-PCR), followed by the genomic sequencing of the VP1/2A junction region of the HAV genome. Sequences of 168 nucleotides were compared and analyzed using the BLAST N, Clustal X and PAUP v. 4.10b programs. All samples were classified as genotype I, with 10 belonging to subgenotype IA and eight to subgenotype IB. The subgenotype IA isolates showed greater diversity than the subgenotype IB isolates at the nucleotide level. Elevated identity values were found between isolates obtained in this study and those from other regions of the world, including Brazil, highlighting the high conservation among different isolates of this virus. However, changes in the HAV subgenotype circulation could also be observed during the evaluated period.
acute hepatitis; nucleotide sequencing; subgenotype IA; subgenotype IB
ARTICLES
Molecular characterization of hepatitis A virus isolates from Goiânia, Goiás, Brazil
Fabíola Souza FiaccadoriI, +; Maristela PereiraII; Alexandre Siqueira Guedes CoelhoIII; Ana Maria Tavares BorgesI; Juliana Alves ParenteII; Célia Maria de Almeida SoaresII; Divina das Dôres de Paula CardosoI
ILaboratório de Virologia, Instituto de Patologia Tropical e Saúde Pública, Instituto de Ciências Biológicas, Universidade Federal de Goiás, Rua 235 Esq. 1ª Av., Setor Universitário, 74.605-050 Goiânia, GO, Brasil
IILaboratório de Biologia Molecular, Instituto de Ciências Biológicas, Universidade Federal de Goiás, Rua 235 Esq. 1ª Av., Setor Universitário, 74.605-050 Goiânia, GO, Brasil
IIILaboratório de Genética e Genômica de Plantas, Instituto de Ciências Biológicas, Universidade Federal de Goiás, Rua 235 Esq. 1ª Av., Setor Universitário, 74.605-050 Goiânia, GO, Brasil
ABSTRACT
Hepatitis A virus (HAV) infection is a public health problem worldwide and the virus has been classified into six genotypes. In Brazil, the only genotype that has been found is genotype I, predominately from subgenotype IA. Here, the HAV genotypes were analyzed of 18 isolates circulating between 1996-2001 in Goiânia, state of Goiás, Brazil. Viral RNA was extracted from 18 serum samples and amplified (RT-PCR/nested-PCR), followed by the genomic sequencing of the VP1/2A junction region of the HAV genome. Sequences of 168 nucleotides were compared and analyzed using the BLAST N, Clustal X and PAUP v. 4.10b programs. All samples were classified as genotype I, with 10 belonging to subgenotype IA and eight to subgenotype IB. The subgenotype IA isolates showed greater diversity than the subgenotype IB isolates at the nucleotide level. Elevated identity values were found between isolates obtained in this study and those from other regions of the world, including Brazil, highlighting the high conservation among different isolates of this virus. However, changes in the HAV subgenotype circulation could also be observed during the evaluated period.
Key words: acute hepatitis - nucleotide sequencing - subgenotype IA - subgenotype IB
Hepatitis A virus (HAV) infection constitutes a public health problem all over the world, including Brazil. HAV is a positive single-stranded RNA virus, classified as the prototype member of the genus Hepatovirus in the family Picornaviridae (ICTVdB 2006). The viral open reading frame (ORF) encodes a precursor polyprotein, which through successive cleavages, gives rise to the structural proteins (VP1-4), which are the viral capsid constituents and the non-structural proteins (2A-C and 3A-D) (Siegl & Frosner 1978, Cohen et al. 1987, Totsuka & Moritsugu 1999).
HAV isolates belong to a single serotype (Lemon & Binn 1983, Lemon & Robertson 1993), but genomic heterogeneity has been demonstrated based on different genome regions, including those encoding for the virion protein VP3 C-terminus (Jansen et al. 1990), the VP1 N-terminus (Robertson et al. 1991, 2000, Costa-Mattioli et al. 2001b) and the VP1/2A junction (Robertson et al. 1992, 2000, Bruisten et al. 2001, Byun et al. 2001, Costa-Mattioli et al. 2001a), which is considered to be the most variable (Robertson et al. 1992).
The characterization of these variants was carried out through phylogenetic analyses, which identified six genotypes (I-VI) that differ among themselves by 15-25%. Genotypes I, II and III were further subdivided into subgenotypes A and B, which differ in approximately 7.5% of the base positions (Robertson et al. 1992, Costa-Mattioli et al. 2002, Lu et al. 2004). Genotype I is predominant all over the world, particularly subgenotype IA (Robertson et al. 1992, Bruisten et al. 2001, Byun et al. 2001, Costa-Mattioli et al. 2001a, b, Hussain et al. 2005, Poovorawan et al. 2005). In Brazil, the only genotype that has been found is genotype I, especially subgenotype IA (de Paula et al. 2004). However, subgenotype IB has also been observed to circulate in the states of Rio de Janeiro and Pernambuco and in the Amazon region (de Paula et al. 2002, 2006, Devalle et al. 2003, Villar et al. 2004, 2006).
In state of Goiás, a genomic characterization study of nine HAV isolates carried out by de Paula et al. (2004) identified only subgenotype IA. The present study, based on samples obtained from 1996-2001, shows the circulation of both IA and IB HAV subgenotypes in different periods of time in Goiânia, state of Goiás, Brazil.
PATIENTS, MATERIAL AND METHODS
Study material - This study was carried out on 18 HAV RNA positive samples, which were obtained from serum samples from individuals with clinical symptoms of viral hepatitis. Thirteen of these samples were anti-HAV-IgM positive. Twelve samples were from males and six from females, from patients ranging in age from 3-40 years old. The study was approved by the Research Ethical Committee of the Federal University of Goiás (number 22000000289).
Detection of HAV RNA- The viral ssRNA was extracted from 400 µL of each serum sample, using the method described by Boom et al. (1990) with modifications (Fiaccadori et al. 2006). Reverse transcription was carried out with the random primer pd(N)6 (Random Hexamer, GE Healthcare, Little Chalfont, UK) following the protocol described by de Paula et al. (2002) with modifications (Fiaccadori et al. 2006).
The amplification reactions, as well as the primers designed for the VP1/2A junction region, followed the protocol described by de Paula et al. (2002). The first amplification reaction was carried out using 5 µL of cDNA. The reaction mix was added for a final volume of 25 µL and contained 0.8 mM of each primer, F06 (CTATTCAGATTGCAAATTAYAAT) and F07 (AAYTTCATYATTTCATGCTCCT). The nested PCR reaction was carried out using 1 µL of the amplicon and the internal primers, F8 (TATTTGTCTGTYACAGAACAATCAG) and F9 (AGGRGGTGGAAGYACTTCATTTGA). The amplicons obtained were separated in a 2% agarose gel stained with ethidium bromide (0.5 µg/mL) for observation of the DNA fragments with the expected size of 244 bp. Milli-Q sterile water and the HAV RNA positive serum sample were used as the reaction controls, with the latter being kindly provided by Instituto Oswaldo Cruz, Rio de Janeiro, RJ, Brazil.
Nucleotide sequencing- Amplicons of the expected size were purified using the comercial kit Qiaquick® PCR Purification (Qiagen, Uniscience, SP, Brazil) following the manufacturer's instructions. Based on the visualization of the DNA fragment in a 0.8% agarose gel stained with ethidium bromide (0.5 µg/mL), DNA quantification was performed by visual comparisons with DNA products with previously known concentration. Products with at least 50-100 ng of DNA/µL of reaction were considered satisfactory for the DNA sequencing procedure.
The direct DNA sequencing reaction was carried out in both directions, using an automatic sequencer (MegaBACE 1000, GE Healthcare) and following the procedure described by Sanger et al. (1977). The reaction system consisted of F08 and F09 primers, Thermo Sequenase enzyme (32 U/µL) (GE Healthcare) and the DYEnamic ET Dye Terminator Cycle Sequencing Kit for MegaBACE DNA Analysis Systems (GE Healthcare).
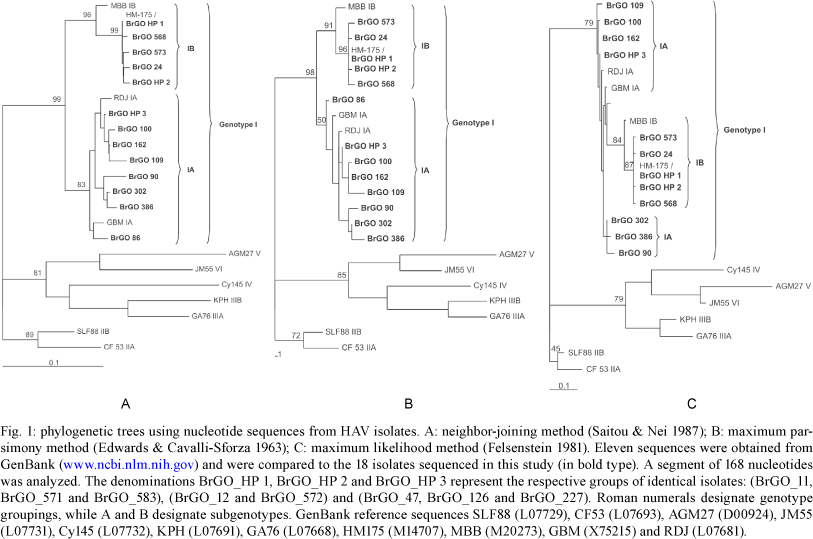
Sequence analysis and phylogenetic relations- The sequences obtained were analyzed initially using the BLAST N program (ICTVdb 2006) (Altschul et al. 1990). For a comparative analysis between the obtained sequences and the previously published HAV sequences (www.ncbi.nlm.nih.gov/genbank/index.html), a region of 168 nucleotides from position 3024-3191 of the VP1/2A junction region of the HAV genome were aligned using the Clustal X program (Thompson et al. 1997). The phylogenetic analysis was performed using neighbor-joining (Saitou & Nei 1987), maximum parsimony (Edwards & Cavalli-Sforza 1963) and maximum likelihood (Felsenstein 1981) methods using the PAUP v.4.10b program (Swofford 1998). Node solidity was evaluated by the bootstrap procedure (Felsenstein 1985). The phylogenetic trees obtained were rooted with the HAV nucleotide sequences representative of genotypes IIA (CF 53), IIB (SLF88), IIIA (GA76), IIIB (KPH), IV (Cy145), V (AGM27) and VI (JM55). The sequences reported in the present study were deposited in the GenBank database under accession numbers EU430063-EU430075.
RESULTS
The comparative analysis of the 18 nucleotide sequences corresponding to a 168 bp region the VP1/2A junction region showed nucleotide identity percentages ranging from 88-100%. In addition, the product of the 18 nucleotide sequences (168 bp) was compared with the corresponding sequence of the prototype strain HM-175 and the percentage of the nucleotide identity also ranged from 88-100% (data not shown).
Furthermore, the comparison of these sequences with different HAV strains previously published and available in the public DNA sequence database (GenBank, ICTVdb 2006) showed high identity (96-100%) with sequences classified as genotype I and particularly with samples isolated in Brazil (Table).
Fig. 1 shows the phylogenetic trees constructed based on the isolates from this study plus previously published reference sequences representative of all six genotypes and their respective subgenotypes (ICTVdb 2006). The trees were rooted with the representative sequences of genotypes IIA, IIB, IIIA, IIIB, IV, V and VI. All of the isolates clustered in genotype I, with eight (44.5%) of these clustered in subgenotype IB and 10 (55.5%) in subgenotype IA, confirming the BLAST N results (data not shown).
Subgenotype IA isolates presented diversity with the percentages of the nucleotide identity ranging from 94-100%. The isolates BrGO_47, BrGO_126 and BrGO_227 were identical to each other and were identified by the denomination BrGO_HP 3 (haplotype 3).
The subgenotype IB isolates presented nucleotide identity percentages ranging from 98-100%, suggesting less diversity within the group. Three isolates, identified by the denomination BrGO_HP 1 (haplotype 1) (BrGO_11, BrGO_571 and BrGO_583) were identical to each other and to the prototype strain HM-175 (M14707). Also, two other isolates, identified by denomination BrGO_HP 2 (haplotype 2) (BrGO_12 and BrGO_572) were identical to each other.
An alternation was observed in the subgenotype circulation during this time period, with the replacement of subgenotype IB at the end of 1996 and its reappearance at the end of 2000 (Table).
The comparison of the predicted amino acid sequence to the VP1/2A junction region of the 18 isolates is shown in Fig. 2. The identity among the predicted sequences ranges from 98-100% among isolates IA and IB. The identity among the isolates from different genotypes ranges from 96-98% (data not shown). In the 25th amino acid position, the IA isolates contain an Arginine (R-Arg) and the IB isolates have a Lysine (K-Lys).
DISCUSSION
The identification of subgenotypes IA and IB in this study corroborates previous studies done in other regions of the country. This demonstrates that a genotype I infection has an endemic character in Brazil, although it is the only South American country in which the circulation of both subgenotypes IA and IB has been reported (de Paula et al. 2002, 2006, Devalle et al. 2003, Villar et al. 2004, 2006). However, this study differs from that of de Paula et al. (2004), in which the analysis of the nucleotide sequence of the VP1/2A junction region showed the occurrence of only subgenotype IA in isolates from the state of Goiás. This would argue for the analysis of a greater number of samples so additional information could be obtained in order to increase the knowledge about HAV strains in the region.
The isolates in this study showed a high degree of identity with other Brazilian strains, which was also observed in a study carried out with isolates from the state of Rio de Janeiro (Villar et al. 2004), suggesting the endemic circulation of isolates throughout the country without the importation of strains from other regions of the world. Additional studies are in progress to better understand the evolutionary character of HAV isolates obtained in different parts of the world.
Although the number of isolates analyzed was small, an alternation in the circulating subgenotypes was observed. Subgenotype IB circulated until the middle of 1996, when it was replaced by IA, which circulated until the beginning of 2000. At this point, IB reemerged and circulated until 2001. To our knowledge, data in the literature showing HAV isolate alternation for comparisons do not exist, so additional studies are necessary in this area of research.
The diversity observed among the different genotype isolates (II, III, IV, V and VI) inferred from the length of the phylogenetic tree branch, was greater than that observed among genotype I isolates. The subgenotype IA isolates differed more from each other in terms of their nucleotide sequences than those belonging to subgenotype IB. From a phylogenetic point of view, there was strong agreement in the results obtained by the three different applied methodologies (neighbor-joining, maximum parsimony and maximum likelihood). However, the results obtained from the maximum likelihood method suggest that the IB subgenotype could have originated from mutations in the subgenotype IA isolates. This hypothesis is corroborated by the lesser diversity found among the IB isolate cluster than the IA isolate cluster. A wider ranged analysis is being performed to investigate this hypothesis.
The predicted amino acid sequences showed a percentage of identity ranging from 98-100% in both subgenotypes (data not shown). Among the substitutions occurring in these sequences, an Arginine (R-Arg) in the IA isolates and a Lysine (K-Lys) in the IB isolates in position 25 of the predicted amino acid sequences stand out. These amino acids both contain positively charged atoms, but it is possible that this substitution does not induce a change in function. This substitution was also observed among isolates in the state of Pernambuco (Villar et al. 2006), which suggests the adaptation of these subgenotypes in Brazil.
The high identity values of the nucleotide sequences and the predicted amino acid sequences among the isolates in this study and from other regions of the world, especially Brazil, emphasize the conservation among the different isolates of this virus. Now that there is an extremely effective vaccine against HAV and currently only a single serotype, there is the possibility of eradicating the disease worldwide if public vaccination campaigns are adopted by the government.
Received 15 August 2008
Accepted 28 November 2008
Financial support: CNPq
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DA 1990. Basic local alignment search tools. J Mol Biol 215: 403-410.
- Boom R, Sol CJA, Salimans MMM, Jansen CL, Werthem-Ven-Dillean PME, Noordaa JVD 1990. Rapid and simple method for purification of nucleic acids. J Clin Microbiol 28: 495-503.
- Bruisten SM, van Steenbergen JE, Pijl AS, Niesters HGM, van Doornum GJJ, Coutinho RA 2001. Molecular epidemiology of hepatitis A virus in Amsterdam, the Netherlands. J Med Virol 63: 88-95.
- Byun KS, Kim JH, Song K-J, Baek LJ, Song J-W, Park SH, Kwon OS, Yeon JE, Kim JS, Bak YT, Lee CH 2001. Molecular studies: hepatitis A virus and hepatectomy. Molecular epidemiology of hepatitis A virus in Korea. J Gastroenterol Hepatol 16: 519-524.
- Cohen JI, Ticehurst JR, Purcell RH, Buckler-White A, Baroudy BM 1987. Complete nucleotide sequence of wild-type hepatitis A virus: comparison with different strains of hepatitis A virus and other picornaviruses. J Virol 61: 50-59.
- Costa-Mattioli M, Cristina J, Romero H, Perez-Bercof R, Casane D, Colina R, Garcia L, Vega I, Glikman G, Romanowsky V, Castello A, Nicand E, Gassin M, Billaudel S, Ferré V 2002. Molecular evolution of hepatitis A virus: a new classification based on the complete VP1 protein. J Virol 76: 9516-9525.
- Costa-Mattioli M, Ferre V, Monpoeho S, Garcia L, Colina R, Billaudel S, Vega I, Perez-Bercoff R, Cristina J 2001a. Genetic variability of hepatitis A virus in South America reveals heterogeneity and co-circulation during epidemic outbreaks. J Gen Virol 82: 2647-2652.
- Costa-Mattioli M, Monpoeho S, Schvoerer C, Besse B, Aleman M-H, Billaudel S, Cristina J, Ferre V 2001b. Genetic analysis of hepatitis A virus outbreak in France confirms the co-circulation of subgenotypes Ia, Ib and reveals a new genetic lineage. J Med Virol 65: 233-240.
- de Paula VS, Baptista ML, Lampe E, Niel C, Gaspar AMC 2002. Characterization of hepatitis A virus isolates from subgenotypes IA and IB in Rio de Janeiro, Brazil. J Med Virol 62: 22-27.
- de Paula VS, Lu L, Niel C, Gaspar AMC, Robertson BH 2004. Genetic analysis of hepatitis A virus isolates from Brazil. J Med Virol 73: 378-383.
- de Paula VS, Niel C, Teves SC, Villar LM, Virgolino H, Gaspar AMC 2006. Molecular epidemiology of hepatitis A virus in Brazilian Amazon. J Gastroenterol Hepatol 21: 1435-1438.
- Devalle S, de Paula VS, Oliveira JM, Niel C, Gaspar AMC 2003. Hepatitis A virus infection in hepatitis C brazilian patients. J Infect 47: 125-128.
- Edwards AWF, Cavalli-Sforza LL 1963. The reconstruction of evolution. Ann Hum Genet 27: 105-106.
- Felsenstein J 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17: 368-376.
- Felsenstein J 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783-791.
- Fiaccadori FS, Soares CMA, Borges AMT, Cardoso DDP 2006. Prevalence of hepatitis A virus infection in Goiânia, Goiás, Brazil, by molecular and serological procedures, 1995-2002. Mem Inst Oswaldo Cruz 101: 423-426.
- Hussain Z, Das BC, Husain SA, Asim M, Chattopadhyay S, Malik A, Poovorawan Y, Theamboonlers A, Kar P 2005. Hepatitis A viral genotypes and clinical relevance: clinical and molecular characterization of hepatitis A virus isolates from Northern India. Hepatol Res 32: 16-24.
- ICTVdb-The Universal Virus Database, version 4. 2006. [cited 2006 Aug 28]. Available from: http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/
- Jansen RW, Siegl G, Lemon SM 1990. Molecular epidemiology of human hepatitis A virus defined by an antigen-capture polymerase chain reaction method. Proc Natl Acad Sci USA 87: 2867-2871.
- Lemon SM, Binn LN 1983. Serum neutralizing antibody response to hepatitis A virus. J Infect Dis 148: 1033-1039.
- Lemon SM, Robertson BH 1993. Current perspectives in the virology and molecular biology of hepatitis A virus. Semin Virol 4: 285-295.
- Lu L, Ching KZ, De Paula VS, Nakano T, Siegl G, Weitz M, Ro-bertson BH 2004. Characterization of the complete genomic sequence of genotype II hepatitis A virus (CF53/Berne isolate). J Gen Virol 85: 2943-2952.
- Poovorawan Y, Theamboonlers A, Chongsrisawat V, Jantaradsamee P, Chutsirimongkol S, Tangkijvanich P 2005. Clinical features and molecular characterization of hepatitis A virus outbreak in a child care center in Thailand. J Clin Virology 32: 24-28.
- Robertson BH, Averhoff F, Cromeans TL, Han X-H, Khoprasert B, Nainan OV, Rosenberg J, Paikoff L, DeBess E, Shapiro CN, Margolis HS 2000. Genetic relatedness of hepatitis A virus isolates during a community-wide outbreak. J Med Virol 62: 144-150.
- Robertson BH, Jansen RW, Khanna B, Totsuka A, Nainan OV, Siegl G, Widell A, Margolis HS, Isomura S, Ito K, Ishizu T, Moritsugu Y, Lemon SM 1992. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J Gen Virol 73: 1365-1377.
- Robertson BH, Khanna B, Nainan OV, Margolis HS 1991. Epidemiologic patterns of wild-type hepatitis A virus determined by genetic variation. J Infect Dis 163: 286-292.
- Saitou N, Nei M 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406-425.
- Sanger F, Nicklen S, Coulson AR 1977. DNA sequencing with chain termination inhibitors. Proc Natl Acad Sci USA 74: 5463-5467.
- Siegl G, Frosner GG 1978. Characterization and classification of virus particles associated with hepatitis A, II. Type and configuration of nucleic acid. J Virol 26: 48-53.
- Swofford DL 1998. PAUP* Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG 1997. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876-4882.
- Totsuka A, Moritsugu Y 1999. Hepatitis A viurs proteins. Intervirology 42: 63-68.
- Villar LM, Lampe E, Meyer A, Gaspar AMC 2004. Genetic variability of hepatitis A virus isolates in Rio de Janeiro: implications for the vaccination of school children. Braz J Med Biol Res 37: 1779-1787.
- Villar LM, Morais LM, Aloise R, Melo MMM, Calado IA, Lampe E, Gaspar AMC 2006. Co-circulation of genotypes IA and IB of hepatitis A virus in Northeast Brazil. Braz J Med Biol Res 39: 873-881.
Publication Dates
-
Publication in this collection
10 Feb 2009 -
Date of issue
Dec 2008
History
-
Received
15 Aug 2008 -
Accepted
28 Nov 2008

 Molecular characterization of hepatitis A virus isolates from Goiânia, Goiás, Brazil
Molecular characterization of hepatitis A virus isolates from Goiânia, Goiás, Brazil