Abstracts
Trypanosoma cruzi (Schyzotrypanum, Chagas, 1909), and Chagas disease are endemic in captive-reared baboons at the Southwest Foundation for Biomedical Research, San Antonio, Texas. We obtained PCR amplification products from DNA extracted from sucking lice collected from the hair and skin of T. cruzi-infected baboons, with specific nested sets of primers for the protozoan kinetoplast DNA, and nuclear DNA. These products were hybridized to their complementary internal sequences. Selected sequences were cloned and sequencing established the presence of T. cruzi nuclear DNA, and minicircle kDNA. Competitive PCR with a kDNA set of primers determined the quantity of approximately 23.9 ± 18.2 T. cruzi per louse. This finding suggests that the louse may be a vector incidentally contributing to the dissemination of T. cruzi infection in the baboon colony.
Baboons; Papio hamadrias; Lice; Pedicinus obtusus; Trypanosoma cruzi infection
As infecções pelo Trypanosoma cruzi e a doença de Chagas são endêmicas em babuínos (Papio hamadryas) reproduzidos em cativeiro na Southwest Foundation for Biomedical Research, em Santo Antonio, Texas. Nós obtivemos produtos de amplificação por PCR do DNA extraído de piolhos colhidos do cabelo e da pele de babuínos chagásicos, com primers aneladores específicos para DNAs nuclear e de cinetoplasto do protozoário. Esses produtos foram hibridizados com suas respectivas seqüências internas complementares. Seqüências selecionadas foram clonadas e o sequenciamento demonstrou a presença de DNA nuclear de T. cruzi, e de minicírculo de kDNA. A PCR competitiva com primers de kDNA determinou a quantidade de aproximadamente 23.9 ± 18.2 T. cruzi por piolho. Este achado sugere que o piolho pode ser um vetor contribuindo para a disseminação de T. cruzi na colônia de babuínos.
BLOOD-SUCKING LICE MAY DISSEMINATE Trypanosoma cruzi INFECTION IN BABOONS
Enrique R. ARGAÑARAZ(1), Gene B. HUBBARD(2), Larissa A. RAMOS(1), Allen L. FORD(3), Nadjar NITZ(1), Michelle M. LELAND(4), John L. VANDEBERG(3) & Antonio R.L. TEIXEIRA(1)
SUMMARY
Trypanosoma cruzi (Schyzotrypanum, Chagas, 1909), and Chagas disease are endemic in captive-reared baboons at the Southwest Foundation for Biomedical Research, San Antonio, Texas. We obtained PCR amplification products from DNA extracted from sucking lice collected from the hair and skin of T. cruzi-infected baboons, with specific nested sets of primers for the protozoan kinetoplast DNA, and nuclear DNA. These products were hybridized to their complementary internal sequences. Selected sequences were cloned and sequencing established the presence of T. cruzi nuclear DNA, and minicircle kDNA. Competitive PCR with a kDNA set of primers determined the quantity of approximately 23.9 ± 18.2 T. cruzi per louse. This finding suggests that the louse may be a vector incidentally contributing to the dissemination of T. cruzi infection in the baboon colony.
KEYWORDS: Baboons; Papio hamadrias; Lice;Pedicinus obtusus; Trypanosoma cruzi infection.
INTRODUCTION
American trypanosomiasis is a zoonanthroponosis caused by the kinetoplastid protozoan Trypanosoma cruzi. The infection leads to Chagas disease, a chronic consumptive ailment affecting the heart and the digestive tube of susceptible hosts25. Wild mammals dwelling on the American continents serve as T. cruzi reservoirs that maintain the parasite in nature12. The geographic distribution of T. cruzi infection coincides with that of its vector, blood-feeding reduviid bugs of the subfamily Triatominae that are found in tropical and subtropical regions, within latitudes 42 °N to 43 °S. When the protozoan infection gets established in the reduviid host it persists for the entire 1-year life of the host. The triatomids are nocturnal, hiding in the daytime and emerging at night when they attack the sleeping prey. The sylvatic species may fly from their diurnal hiding places to feed on a host located some distance away22. Contamination of a host with metacyclic forms of the parasite in the feces of the reduviids has been associated with the human disease25. Serological evidences have shown the prevalence of T. cruzi infections ranges from 0.43 per cent in Georgia, USA7, to 74.2 per cent in rural areas of Santiago del Estero, Argentina6. It has been estimated that 18 million people are chronically infected with T. cruzi, and 35 million are at risk of contracting the infection31.
Transplacental transmission of T. cruzi may occur in 2.5% of human fetuses of infected mothers2,3,9,20. Breast-feeding has been associated with transmission of T. cruzi from infected mothers to their infants, but further studies are needed to determine the epidemiologic importance of this route of transmission15. T. cruzi has been considered the most infective blood protozoan, and several cases of accidental transmission to laboratory workers have been reported4. In addition, oral transmission of T. cruzi is well documented, and is often related to consumption of insectivorous and carnivorous mammals25.
The Anoplura (or sucking lice) live on the surface of the host. To our knowledge, the possibility that sucking lice of the family Pediculidae Linaeus, 1758, comprising the genera Pedicinus, Pediculus and Phthirus, are capable of transmiting of T. cruzi has not previously been investigated. We have tested this hypothesis in a baboon (Papio hamadryas, Cercopithecidae) colony16, which is maintained in large open-air pens in San Antonio, Texas19. In this colony, serological data indicate T. cruzi infection in 9.4% of 2-to-3-year-olds, 14.0% of 7-to-10-year-olds, and 22.5% of baboons that are 15 years old or older (unpublished data). In this study we show that sucking lice, Pedicinus obtusus, captured from nine baboons reared in the colony, yielded PCR amplification products of T. cruzi kinetoplast (kDNA) and nuclear (nDNA) DNA with specific primers. Cloning and sequencing the PCR products confirmed the presence of the parasite DNA in the lice.
MATERIALS AND METHODS
Baboons: This investigation was conducted with animals from a breeding colony of approximately 3,300 baboons, whose members have been useful animal models in many areas of biomedical research29,30. An earlier observation of natural death of an infant baboon8 infected with T. cruziand a more recent observation of three baboons that died spontaneously and exhibited nests of amastigote forms of T. cruzi in host cells and lesions consistent with Chagas heart disease (unpublished data) prompted us to search T. cruzi infection in a cohort of 30 baboons. The enzyme-linked immunosorbent assay, indirect hemagglutination, and indirect immunofluorescence assays28 were used to determine the presence of specific antibodies in the serum. These positive assays are regarded as evidence of T. cruzi infection31. In adition, 2 mL blood samples drawn from six baboons were seeded in 10 mL of liver infusion tryptose medium. Aliquots of each culture tube were examined monthly for eight months.
Collection of lice and extraction of DNA: We collected P. obtusus (Fig. 1) from the hair and skin of severely parasitized baboons in the colony. Bulk samples of lice from each baboon was pooled, fixed in 70% ethanol and subjected to DNA extraction1,21. DNA was extracted from the blood, and from the heart of baboons that died of Chagas disease. DNA was also extracted from epimastigote forms of Berenice T. cruzi grown in axenic medium27 and from macrophage line P388DI-IL1 grown in Dulbecco's Minimum Essential Medium supplemented with 20% fetal calf serum, 100 mg/mL streptomycin and 100 IU penicillin26.
Polymerase chain reaction (PCR): DNA was analyzed by PCR with specific primers for the constant minicircle kDNA region1,21,23, and for highly repetitive sequence18 of nDNA of T. cruzi. The kDNA primer set (S35, 5'-ATAATGTACGGG(T/G)GAGATGC-3' and S36, 5'-GGTTCGATTGGGGTTGGTG-3') annealing to the constant region of minicircles yields a 330 bp product and its catamer of 660 bp23. The nDNA primer set (PON1, 5'-TGGCTTGGAGGAGTTATTGT-3' and PON2, 5'-AGGAGTGACGGTTGATCAGT-3') amplifies a 250 bp fragment18. A DNA thermal cycler (MJ Research, Watertown, MA) was used for 30-32 cycles as follows: PON1/2, 94 °C for 2 min, 58 °C for 1 min, and 72 °C for 1 min; S35/36, 94 °C for 2 min, 64 °C for 1 min, and 72 °C for 1 min. The reactions were run with 100 ng of lice target template and with 50 pg of DNA from T. cruzi culture forms. Each reaction was done in 25 mL aliquots containing 2.5 U of Taq polymerase (Perkin Elmer, Cetus Norwalk), 0.2 mM dNTPs, 50 mM Tris-HCl (pH 9.0 at 20 °C), 1.5 mM MgCl2, 200 mM ammonium sulfate, and 5 mM each primer. A 10 mL aliquot from each PCR reaction was subjected to a 1% agarose gel eletrophoresis. Negative controls (water, healthy baboon DNA, and P388D1-IL1 DNA) and positive control (T. cruzi DNA) were always included to detect DNA contamination and ensure that the PCR worked efficiently.
Competition assays: After the presence of T. cruzi in sucking lice was confirmed in nine bulk samples, we ran competitive PCR assays with S35/36 primers set to quantify the number of parasites in these samples5. These assays consisted of a mixture of an unknown quantity of T. cruzi template minicircle DNA with serial dilutions of a known quantity of competitor DNA (pT7 Blue vector containing the 280 pb kDNA fragment). The competitor fragment binds the same primers and yields a product that is distinguishable from the sample template (280 versus. 330 bp). The equivalency point corresponds to the tube with equimolar concentrations of template and competitor PCR products in the reaction mix. The determination of equivalency points was made by visual comparison in 1.5% wide range agarose/0.5% standard agarose gel. In all PCR positive samples, using the known amount of the competitor DNA in the reaction, the unknown quantity of the template was calculated, under the asssumption that there were 10,000 minicircles/parasite and, thus 15 fg of 330 bp template/parasite5.
Hybridization, cloning and sequencing: The PCR amplification products were transferred by capillarity to a nylon membrane. Prehybridization and hybridization were performed in 6X SSC, 5X Denhardt's, 0.5% SDS and 100 µg/mL salmon DNA solution. The membranes were prehybridized for 4 hr, and then were hybridized for 12 hr with internal oligonucleotide sequences for different amplified fragments. For S35/36 products23, the sequence was S67 5'-GGTTTTGGGAGGGG(CG)-(G/C)-(T/G)TC-3'. For PON1/2 products18, the sequence was PON3 5'-CCGGCCTGTGTCTGCGGC-3'. These oligonucleotides were radiolabelled with [g 32P]-dATP (3000 CimMol) using the polynucleotide kinase method following the manufacturer's recomendation (Life Technologies). After the membrane was washed once for 5 min with 1X SSC and 0.1% SDS and twice for 30 min each with 0.2X SSC and 0.1% SDS, it was autoradiographed for variable periods of time.
The amplification products obtained with specific kDNA primers were cloned into PCRII vector, and several clones were selected and subjected to sequencing using an f-mol DNA cycle sequencing system (Promega). The sequences were submitted to PDU, GBU, GenBank and EMBL blast analyses.
RESULTS
Phenotypic and genotypic markers of Trypanosoma cruzi infection: Phenotypic and genotypic markers that indicate the presence of T. cruzi infection in the baboon colony reared in outdoor cages at the Southwest Foundation for Biomedical Research, San Antonio, Texas, are shown in Table 1. The results of three serologic assays covalidated the PCR amplification products from target template T. cruzi DNA extracted from blood or tissue samples of 30 baboons. Furthermore, the culture of blood from three out of six baboons yielded parasitic forms of T. cruzi that were inoculated in BALB/c mice. Culture of macerated louse tissue was not performed because samples were received in 70% ethanol. The animals showed trypomastigote forms in the blood and amastigote forms of T. cruzi encysted in tissue cells (data not shown). Then, we collected lice from the skin of those three baboons that showed parasitemia and from six others showing positive serologic tests and clinical evidence of the infection.
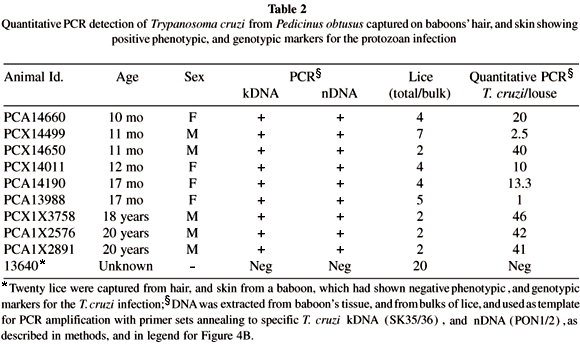
Detection of Trypanosoma cruzi from Pedicinus obtusus: Here we tested the hypothesis that the blood sucking lice P. obtusus may play some role in disseminating the T. cruzi among members of the baboon colony. In the absence of direct demonstration of live T. cruzi in lice, the evidence supporting this hypothesis is derived from PCR amplification of sequences from lice target template DNA with specific nested sets of T. cruzi kDNA23, and nDNA18 primers. Table 2 summarizes results of experiments showing that lice captured from the hair, and from the skin of baboons with Chagas disease are infected with the T. cruzi. This evidence was given by PCR with nested sets of primers annealing to the protozoan kDNA, and nDNA templates. In the control experiment, 20 lice collected from the hair, and from the skin from a healthy baboon reared in a cage far apart from that having T. cruzi infected baboons, and that yielded negative PCR for the protozoan infection.
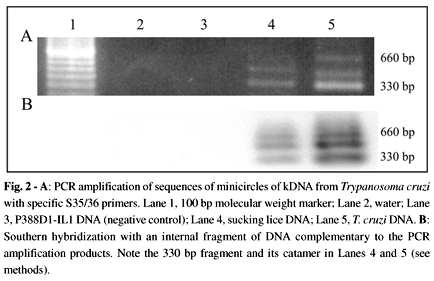
Figure 2A shows PCR amplification products with specific kDNA primers. The amplification products hybridized with an internal kDNA probe (Fig. 2B). Sequencing of the PCR amplification product showed typical constant and hypervariable kDNA minicircle regions. This sequence is deposited in GenBank (accession number AF114153). Confirmation of the presence of T. cruzi DNA in the target template extracted from bulk samples of lice (Table 2) was obtained by PCR with a PON1/2 nested set of primers (Fig. 3A). The amplification product of 250 bp hybridized with its complementary internal sequence, thus showing its specificity (Fig. 3B). This amplification of the parasite nDNA suggested the presence of living T. cruzi in the sucking louse.
Quantitation of living Trypanosoma cruzi in lice: Therefore, we performed competitive, quantitative PCR with a kDNA-nested set of primers to determine the number of parasites in each louse5. To determine the sensitivity of this quantitative procedure we used concentrations of T. cruzi DNA corresponding to 1, 10 and 100 parasites. Figure 4A shows the result of a standard competitive PCR to detect the DNA equivalent of a single parasite. Then, we determined the quantity of parasite DNA in 100 ng of DNA extracted from bulk samples of lice. Interestingly, the quantity of 23.9 ± 18.2 T. cruzi per louse was found. A typical experiment carried out with lice captured from the hair, and from the skin of a 17 month old baboon (Id PCA13988, Table 2) is shown in Figure 4B. The zone of equivalency corresponded to 1.5 fg of competitor DNA. Because this quantity of DNA corresponds to 1/10 parasite5, and because the reaction contained 1/50 of the DNA sample, we calculate that there were approximately five T. cruzi in this bulk sample of five lice tested.
DISCUSSION
We have demonstrated for the first time the contamination of blood-sucking lice of the family Pediculidae with T. cruzi. However, the only evidence of the protozoan in blood sucking lice was obtained by PCR amplification of T. cruzi kDNA and nDNA sequences in target templates with specific nested set of primers18, 23. We believe that the sequence of a minicircle kDNA belongs to a living T. cruzi, instead of being simply kDNA integrated in the genome of blood cells26 sucked in by the louse, because nDNA sequence was also amplified from the lice target template and there is no published evidence that T. cruzi nDNA integrates in the genome of a host. These findings suggest, therefore, that sucking lice are capable of transmiting living T. cruzi infection among baboons, either by contamination of mucosal surfaces, conjunctiva and skin wounds with its intestinal waste, or by the oral route.
The insects belonging to the family Pediculidae are host specific. They are usually transmitted by direct contact. Of interest, it has been shown that allo grooming by Old World monkeys is an altruistic behavior that removes external parasites from others. In particular, grooming of infants by mothers, which appears to be an important component of maternal care, would contribute to the dissemination of the T. cruzi infection, because the mothers pick up lice and eggs of lice and eat them24. It has been reported that insectivorous and carnivorous mammals become contaminated with T. cruzi through the oral route25. The demonstration of T. cruzi kDNA and nDNA in lice captured from baboons supports the hypothesis that contamination of nonhuman primates with the protozoan may contribute to the endemicity of Chagas disease in the colony. The estimated number of T. cruzi in blood sucking lice (23.9 ± 18.2 parasite per louse) suggests that this protozoan does not multiply in the intestine of the bug. However, dissemination of the infection by lice contaminated with T. cruzi is highly plausible, because the infection can be readly acquired through the oral route13,14,17. In this regard, it has been shown that reducing the number of the parasites in the inocula may retard but does not prevent the late course of experimental Chagas disease13. A single T. cruzi trypomastigote form is sufficient to establish an infection to produce widespread, fatal disease14. Although mechanical transmission of T. cruzi has been considered by several authors4,8,13-15,17, its epidemiological importance remains to be determined.
The sucking lice that occur on human beings comprise the species Pediculus humanus, the body louse and head louse, and Phitirus pubis, the crab louse. These lice occur only on man and not on other hosts. The demonstration of PCR amplification products of T. cruzi sequences, with specific nDNA18, and kDNA23 primers, from template DNA obtained from sucking lice of the suborder Anoplura, which infest nonhuman primates, suggests that other sucking lice (P. humanus, and P. pubis) also may transmit the infection. However, the hypothetical epidemiological importance of sucking lice in the dissemination of the T. cruzi infections in human beings may be difficult to demonstrate, in the absence of allo grooming and ingestion of external parasites.
The primary vector for transmission of T. cruzi in this outdoor, baboon colony in San Antonio, TX, is presumed to be hemiptera, reduviid bugs of the family Triatominae. Several species of triatomids found in Southern United States (Triatoma rubrofasciata, T. sanguesuga) have been related to transmission of Chagas disease22. These species have been implicated in an outbreak of acute Chagas disease in a Rhesus monkey colony at Brooks Air Force Base, also in San Antonio10. Interestingly, baboon caretakers and researchers carrying on long term scientific work with baboons are aware that these flying insects occasionally are seen at night at the outdoor cages.
We believe that the T. cruzi infections in the colony were initiated by reduviid bugs, which possibly continued to transmit them, regardless of the lack of information with respect to the presence of the main vector of Chagas disease in the baboon colony. However, our results suggest that T. cruzi infection may be further disseminated in the colony by sucking lice of the suborder Anoplura. We propose that the blood-sucking lice P. obtusus infesting Old World monkeys do transmit T. cruzi infection, as indicated by the presence of specific kDNA and nDNA sequences, probably acquired during feeding on an infected baboon.
RESUMO
Piolhos hematófagos podem disseminar infecção pelo Trypanosoma cruzi em babuínos
As infecções pelo Trypanosoma cruzi e a doença de Chagas são endêmicas em babuínos (Papio hamadryas) reproduzidos em cativeiro na Southwest Foundation for Biomedical Research, em Santo Antonio, Texas. Nós obtivemos produtos de amplificação por PCR do DNA extraído de piolhos colhidos do cabelo e da pele de babuínos chagásicos, com primers aneladores específicos para DNAs nuclear e de cinetoplasto do protozoário. Esses produtos foram hibridizados com suas respectivas seqüências internas complementares. Seqüências selecionadas foram clonadas e o sequenciamento demonstrou a presença de DNA nuclear de T. cruzi, e de minicírculo de kDNA. A PCR competitiva com primers de kDNA determinou a quantidade de aproximadamente 23.9 ± 18.2 T. cruzi por piolho. Este achado sugere que o piolho pode ser um vetor contribuindo para a disseminação de T. cruzi na colônia de babuínos.
ACKNOWLEDGMENTS
We acknowledge Dr. W. C. Van Voorhis, Division of Allergy and Infectious Diseases, SJ-10, Dept. of Medicine, University of Washington, Seattle, USA, for donation of the pT7 Blue vector containing the 280 bp competitor DNA. We thank Ana de Cássia Vexenat, Marie Silva and Sherrian McAnn for technical support. This work was supported in part by NIH grant P51 RR13986 to the Southwest Regional Primate Research Center, and by a grant from the Inter-American Developing Bank/Financiadora de Estudos e Projetos to the Chagas Disease Multidisciplinary Research Laboratory, Faculty of Medicine of the University of Brasilia, Brazil.
Received: 09 March 2001
Accepted: 30 May 2001
Correspondence to: Antonio R.L. Teixeira, Chagas Disease Multidisciplinary Research Laboratory, University of Brasília. P.O. Box 04536, 70919-970 Brasília, Brazil. E-mail: ateixeir@unb.br.
Fax: 55+61 273-4645. Tel.: 55+61 349-4987.
References
- 1. AVILA, H.A.; SIGMAN, D.S.; COHEN, L.M.; MILLIKAN, R.C. & SIMPSON, L. - Polymerase chain reaction amplification of Trypanosoma cruzi kinetoplast minicircle DNA isolated from whole blood lysates: diagnosis of chronic Chagas disease. Molec. Biochem. Parasit., 48: 211-221, 1991.
- 2. AZOGUE, E.; LA FUENTE, C. & DARRAS, C. - Congenital Chagas disease in Bolivia: epidemiological aspects and pathological findings. Trans. roy. Soc. trop. Med. Hyg., 79: 176-180, 1985.
- 3. BITTENCOURT, A.L. - Congenital Chagas disease. Amer. J. Dis. Child., 130: 97-103, 1976.
- 4. BRENER, Z. - Laboratory acquired Chagas disease: an endemic disease among parasitologists? In: MOREL, C.M., ed. Genes and antigens of parasites: a laboratory manual. Rio de Janeiro, Fundaçăo Instituto Oswaldo Cruz, 1984. p. 3-9.
- 5. CENTURION-LARA, A.; BARRETT, L. & VAN VOORTHIS, W.C. - Quantitation of parasitemia by competitive polymerase chain reaction amplification of parasite kDNA minicircles during chronic infection with Trypanosoma cruzi. J. infect. Dis., 170: 1334-1339, 1994.
- 6. CHICHERO, J.A.; BONET, A.H. & SANTAMARÍA, N. - Estudio de la prevalencia de la enfermedad de Chagas en varones de 20 ańos de edad en la Provincia de Santiago del Estero. Sem. méd. (B. Aires), 133: 1792-1798, 1967.
- 7. FARRAR Jr., W.E.; KAGAN, I.G.; EVERTON, F.D. & SELLERS Jr., T.F. - Serologic evidence of human infection with Trypanosoma cruzi in Georgia. Amer. J. Hyg., 78: 166-170, 1963.
- 8. GLEISER, C.A.; YAEGER, R.G. & GHIDONI, J.J. - Trypanosoma cruzi infection in a colony-born baboon. J. Amer. vet. med. Ass., 189: 1225-1226, 1986.
- 9. HOWARD, J.E. & RUBIO, M. - Enfermedad de Chagas congénita. I. Estudio clínico y epidemiológico de 30 casos. Bol. chile. Parasit., 23: 107-112, 1968.
- 10. KASA, T.J.; LATHROP, G.D.; DUPUY, H.J.; BONNEY, C.H. & TOFT, J.D. - An endemic focus of Trypanosoma cruzi infection in a subhuman primate research colony. J. Amer. vet. med. Ass., 171: 850-854, 1977.
- 12. LAURIA-PIRES, L. & TEIXEIRA, A.R.L. - Virulence and pathogenicity associated with diversity of Trypanosoma cruzi stocks and clones derived from Chagas disease patients. Amer. J. trop. Med. Hyg., 55: 304-310, 1996.
- 13. MARSDEN, P.D. - Trypanosoma cruzi infections in CFI mice. I. Mortality with different doses of trypanosomes. Ann. trop. Med. Parasit., 61: 57-61, 1967a.
- 14. MARSDEN, P.D. - Trypanosoma cruzi infections in CFI mice. II. Infections induced by different routes. Ann. trop. Med. Parasit., 61: 62-67, 1967b.
- 15. MILES, M.A. - Trypanosoma cruzi: milk transmission of infection and immunity from mother to young. Parasitology, 65: 1-9, 1972.
- 16. PAGE, S.L.; CHIU, C. & GOODMAN, M. - Molecular phylogeny of Old World monkeys (Cercopithecidae) as inferred from gamma-globin DNA sequences. Molec. Phylogenet. Evol., 13: 348-359, 1999.
- 17. PHILLIPS, N.R. - Experimental studies on the quantitative transmission of Trypanosoma cruzi: considerations regarding the standardization of materials. Ann. trop. Med. Parasit., 54: 60-70, 1960.
- 18. REQUENA, J.M.; JIMENEZ-RUIZ, A.; SOTO, M.; LOPEZ, M.C. & ALONSO, C. - Characterization of a highly repeated interspersed DNA sequence of Trypanosoma cruzi: its potential use in diagnosis and strain classification. Molec. Biochem. Parasit., 51: 271-280, 1992.
- 19. ROGERS, J.; MAHANEY, M.C.; WITTE, S.M. et al - A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatellite polymorphisms. Genomics, 67: 237-247, 2000.
- 20. SCHMUŃIS, G.A. & SZARFMAN, A. - La enfermedad de Chagas congénita. Medicina (B. Aires), 37: 47-53, 1977.
- 21. SIMPSON, L. - The mitochondrial genome of kinetoplastid protozoa: genomic organization, transcription, replication, and evolution. Ann. Rev. Microbiol., 41: 363-382, 1987.
- 22. SJOGREN, R.D. & RYCKMAN, R.E. - Epizootiology of Trypanosoma cruzi in southwestern North America. 8. Nocturnal flights of Triatoma protacta (Uhler) as indicated by collections at black light traps (Hemiptera: Reduvidae: Triatominae). J. med. Ent., 3: 81-92, 1966.
- 23. STURM, N.R.; DEGRAVE, W.; MOREL, C.M. & SIMPSON, L. - Sensitive detection and schizodeme classification of Trypanosoma cruzi cells by amplification of kinetoplast minicircle DNA sequences: use in the diagnosis of Chagas disease. Molec. Biochem. Parasit., 33: 205-214, 1989.
- 24. TANAKA, I. & TAKEFUSHI, H. - Elimination of external parasites (Lice) is the primary function of grooming in free-ranging Japanese macaques. Anthropol. Sci., 101: 187-193, 1993.
- 25. TEIXEIRA, A.R.L. - The Stercorarian Trypanosomes. In: SOULSBY, E.J.L., ed. Immune responses in parasitic infections: Immunology, Immunopathology and Immunoprophylaxis Boca Raton, CRC Press, 1987. p. 25-115.
- 26. TEIXEIRA, A.R.L.; ARGAŃARAZ, E.R.; FREITAS Jr., L.H. et al - Possible integration of Trypanosoma cruzi kDNA minicircles into the host cell genome by infection. Mutat. Res., 305: 197-209, 1994.
- 27. TEIXEIRA, A.R.L.; FIGUEIREDO, F. & REZENDE FILHO, J. - Delayed hypersensitivity to Trypanosoma cruzi antigens. II. Use of the skin test with the T12E antigen for the diagnosis of Chagas disease. Rev. Soc. bras. Med. trop., 28: 259-265, 1995.
- 28. TINOCO, D.L.; GARCIA, M.P.; LAURIA-PIRES, L.; SANTANA, J.M. & TEIXEIRA, A.R. - The use of four immunological exams for the determination of Chagas disease prevalence in streetsweepers of the City Sanitation Service in the Federal District. Rev. Soc. bras. Med. trop., 29: 33-40, 1996.
- 29. TOFT, J.D. & EBERHARD, M.L. - Parasitic diseases. In: BENNET, B.T.; ABEE, C.R. & HENRICKSON, R., ed. Nonhuman primates in biomedical research New York, Academic Press, 1998. p. 115-116.
- 30. VANDEBERG, J.L. & WILLIAMS-BLANGERO, S. - Strategies for using nonhuman primates in genetic research on multifactorial diseases. Lab. Anim. Sci., 46: 146-151, 1996.
- 31. WORLD HEALTH ORGANIZATION - Control of Chagas disease. Report of a WHO Expert Committee. Wld. Hlth. Org. techn. Rep. Ser., (811): 1-95, 1991.
Publication Dates
-
Publication in this collection
31 Oct 2001 -
Date of issue
Oct 2001
History
-
Accepted
30 May 2001 -
Received
09 Mar 2001

 Blood-sucking lice may disseminate Trypanosoma cruzi infection in baboons
Blood-sucking lice may disseminate Trypanosoma cruzi infection in baboons