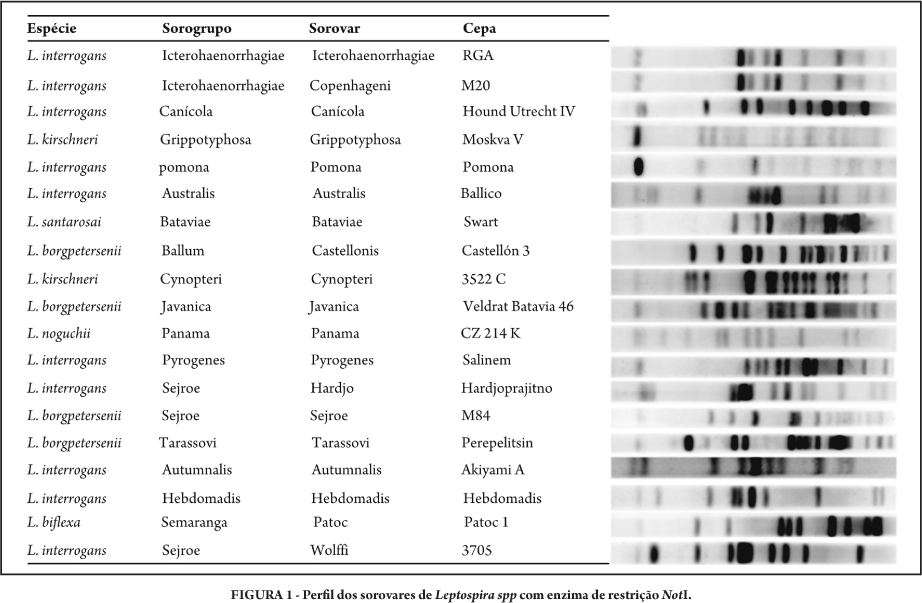
INTRODUCTION: Leptospirosis is an endemic zoonosis of worldwide distribution, caused by bacteria of the genus Leptospira. This genus includes pathogenic and saprophytic species, with more than 200 different serovars, thus making it difficult to characterize. The technique of pulsed field gel electrophoresis has been used as a tool to aid in this characterization. The aims of this study were to standardize the PFGE technique, determine the molecular profiles of reference strains used at the National Reference Laboratory for Leptospirosis/World Health Organization Collaborating Center for Leptospirosis and create a database with these profiles. METHODS: Nineteen strains were analyzed by means of PFGE, using the restriction enzyme NotI. RESULTS: Each strain presented a unique profile that could be considered to be a specific genomic identity, with the exception of the serovars Icterohaemorrhagiae and Copenhageni, whose profiles were indistinguishable. CONCLUSIONS: It was possible to create a database of molecular profiles, which are being used in the Laboratory for comparing and identifying strains isolated from clinical cases.
Leptospira sp; Pulsed Field Gel Electrophoresis; Leptospirosis; Molecular biology

 Characterization of Leptospira sp reference strains using the pulsed field gel electrophoresis technique
Characterization of Leptospira sp reference strains using the pulsed field gel electrophoresis technique